[English] 日本語
 Yorodumi
Yorodumi- EMDB-6370: 3D-Structure of negatively stained Schistosome myosin filament ob... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-6370 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | 3D-Structure of negatively stained Schistosome myosin filament obtained by low-dose electron microscopy | |||||||||
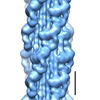
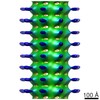
 Map data Map data | Map of Schistosome thick filaments. Initial view is from the Z-line perspective. If the map is rotated by 90 degrees in x direction, the J motif of the interacting heads is featured and the backbone subfilaments can be seen clearly. | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Schistosoma mansoni / rigid docking / single particle reconstruction / Iterative Helical Real Space Reconstruction (IHRSR) / negative stain / thick filament / smooth muscle | |||||||||
| Function / homology |  Function and homology information Function and homology informationmyosin complex / myofibril / cytoskeletal motor activity / actin filament binding / calcium ion binding / ATP binding Similarity search - Function | |||||||||
| Biological species |  | |||||||||
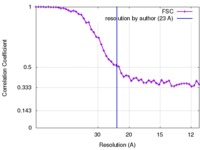
| Method | helical reconstruction / negative staining / Resolution: 23.0 Å | |||||||||
 Authors Authors | Sulbaran G / Alamo L / Pinto A / Marquez G / Mendez F / Padron R / Craig R | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2015 Journal: Proc Natl Acad Sci U S A / Year: 2015Title: An invertebrate smooth muscle with striated muscle myosin filaments. Authors: Guidenn Sulbarán / Lorenzo Alamo / Antonio Pinto / Gustavo Márquez / Franklin Méndez / Raúl Padrón / Roger Craig /   Abstract: Muscle tissues are classically divided into two major types, depending on the presence or absence of striations. In striated muscles, the actin filaments are anchored at Z-lines and the myosin and ...Muscle tissues are classically divided into two major types, depending on the presence or absence of striations. In striated muscles, the actin filaments are anchored at Z-lines and the myosin and actin filaments are in register, whereas in smooth muscles, the actin filaments are attached to dense bodies and the myosin and actin filaments are out of register. The structure of the filaments in smooth muscles is also different from that in striated muscles. Here we have studied the structure of myosin filaments from the smooth muscles of the human parasite Schistosoma mansoni. We find, surprisingly, that they are indistinguishable from those in an arthropod striated muscle. This structural similarity is supported by sequence comparison between the schistosome myosin II heavy chain and known striated muscle myosins. In contrast, the actin filaments of schistosomes are similar to those of smooth muscles, lacking troponin-dependent regulation. We conclude that schistosome muscles are hybrids, containing striated muscle-like myosin filaments and smooth muscle-like actin filaments in a smooth muscle architecture. This surprising finding has broad significance for understanding how muscles are built and how they evolved, and challenges the paradigm that smooth and striated muscles always have distinctly different components. #1:  Journal: BIOPHYS.J. / Year: 2014 Journal: BIOPHYS.J. / Year: 2014Title: Schistosome Muscles Contain Striated Muscle-Like Myosin Filaments in a Smooth Muscle-Like Architecture Authors: Sulbaran G / Alamo L / Pinto A / Marquez G / Mendez F / Padron R / Craig R | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_6370.map.gz emd_6370.map.gz | 1.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-6370-v30.xml emd-6370-v30.xml emd-6370.xml emd-6370.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_6370_fsc.xml emd_6370_fsc.xml | 5.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_6370.tif emd_6370.tif emd_6370_1.tif emd_6370_1.tif emd_6370_2.tif emd_6370_2.tif | 267.7 KB 285.8 KB 237.1 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-6370 http://ftp.pdbj.org/pub/emdb/structures/EMD-6370 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6370 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-6370 | HTTPS FTP |
-Validation report
| Summary document |  emd_6370_validation.pdf.gz emd_6370_validation.pdf.gz | 333.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_6370_full_validation.pdf.gz emd_6370_full_validation.pdf.gz | 332.8 KB | Display | |
| Data in XML |  emd_6370_validation.xml.gz emd_6370_validation.xml.gz | 8.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6370 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6370 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6370 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-6370 | HTTPS FTP |
-Related structure data
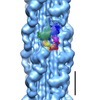
| Related structure data |  3jaxMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_6370.map.gz / Format: CCP4 / Size: 8.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_6370.map.gz / Format: CCP4 / Size: 8.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of Schistosome thick filaments. Initial view is from the Z-line perspective. If the map is rotated by 90 degrees in x direction, the J motif of the interacting heads is featured and the backbone subfilaments can be seen clearly. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 5.7 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Myosin thick filaments from Schistosoma mansoni smooth muscle
| Entire | Name: Myosin thick filaments from Schistosoma mansoni smooth muscle |
|---|---|
| Components |
|
-Supramolecule #1000: Myosin thick filaments from Schistosoma mansoni smooth muscle
| Supramolecule | Name: Myosin thick filaments from Schistosoma mansoni smooth muscle type: sample / ID: 1000 Details: S. mansoni relaxed myosin thick filaments were isolated by permeabilizing and homogenizing whole animals in relaxing solution, centrifuged, and resuspended in blebbistatin. Oligomeric state: polymer of myosin II molecules helically assembled over a paramyosin core Number unique components: 1 |
|---|
-Macromolecule #1: Myosin II
| Macromolecule | Name: Myosin II / type: protein_or_peptide / ID: 1 / Name.synonym: Myosin Type II Details: Myosin II is a protein complex formed by two heavy chains and two associated light chains (for each myosin head), plus additional proteins such as paramyosin. Using ATP hydrolysis, myosin ...Details: Myosin II is a protein complex formed by two heavy chains and two associated light chains (for each myosin head), plus additional proteins such as paramyosin. Using ATP hydrolysis, myosin functions as a molecular motor, producing movement by causing actin filaments to slide. Number of copies: 1 Oligomeric state: polymer of myosin II molecules helically assembled over a paramyosin core Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism:  |
| Sequence | UniProtKB: Myosin heavy chain GO: myosin complex, cytoskeletal motor activity, ATP binding InterPro: Myosin, N-terminal, SH3-like, P-loop containing nucleoside triphosphate hydrolase, Myosin head, motor domain, INTERPRO: IPR027401, Myosin tail, IQ motif, EF-hand binding site |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7 Details: 100 mM NaCl, 3 mM MgCl2, 1 mM EGTA, 5 mM PIPES, 1mM NaN3, 5 mM MgATP, 0.01 mM blebbistatin, protease inhibitor cocktail (Sigma P-8465) |
|---|---|
| Staining | Type: NEGATIVE Details: One drop of filament suspension was placed on grids and negatively stained with 1% uranyl acetate. |
| Grid | Details: 400-mesh holey carbon grids. Specimens were imaged on thin carbon extending over the holes. |
| Vitrification | Cryogen name: NONE / Instrument: OTHER |
- Electron microscopy
Electron microscopy
| Microscope | FEI/PHILIPS CM120T |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 240,000 times magnification |
| Details | 1.5 post-magnification, low-dose conditions |
| Date | Mar 1, 2013 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F224 (2k x 2k) / Number real images: 263 / Average electron dose: 10 e/Å2 Details: Images were acquired with a 2K x 2K CCD TVIPS camera model F224HD at 5.7 A/pixel. Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 80 kV / Electron source: LAB6 |
| Electron optics | Calibrated magnification: 42000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.0 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 42000 |
| Sample stage | Specimen holder: Room temperature holder / Specimen holder model: SIDE ENTRY, EUCENTRIC |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | PDB ID: Chain - #0 - Chain ID: A / Chain - #1 - Chain ID: B / Chain - #2 - Chain ID: C / Chain - #3 - Chain ID: D / Chain - #4 - Chain ID: E / Chain - #5 - Chain ID: F |
|---|---|
| Software | Name:  Chimera Chimera |
| Details | 3DTP was fitted as a rigid body using the "Fit in Map" tool of UCSF Chimera. |
| Refinement | Space: REAL / Protocol: RIGID BODY FIT |
| Output model |  PDB-3jax: |
 Movie
Movie Controller
Controller