+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | IAA bound mTAUT | |||||||||||||||
 Map data Map data | ||||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Transporter / Taurine / Chlorine / Sodium / MEMBRANE PROTEIN/IMMUNE SYSTEM / MEMBRANE PROTEIN-IMMUNE SYSTEM complex | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationNa+/Cl- dependent neurotransmitter transporters / Amino acid transport across the plasma membrane / alanine transmembrane transporter activity / taurine:sodium symporter activity / gamma-aminobutyric acid:sodium:chloride symporter activity / amino acid:sodium symporter activity / amino acid import across plasma membrane / taurine transmembrane transport / neurotransmitter transport / microvillus membrane ...Na+/Cl- dependent neurotransmitter transporters / Amino acid transport across the plasma membrane / alanine transmembrane transporter activity / taurine:sodium symporter activity / gamma-aminobutyric acid:sodium:chloride symporter activity / amino acid:sodium symporter activity / amino acid import across plasma membrane / taurine transmembrane transport / neurotransmitter transport / microvillus membrane / positive regulation of cell differentiation / modulation of chemical synaptic transmission / GABA-ergic synapse / basolateral plasma membrane / postsynaptic membrane / apical plasma membrane / plasma membrane Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||||||||
 Authors Authors | She J / Wang M / He J | |||||||||||||||
| Funding support |  China, 4 items China, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2025 Journal: Proc Natl Acad Sci U S A / Year: 2025Title: Molecular basis for substrate recognition and transport of mammalian taurine transporters. Authors: Mingxing Wang / Jin He / Qianwen Cai / Shen-Ao Zhang / Ji She /  Abstract: The taurine transporter (TAUT) mediates cellular taurine uptake, playing a critical role in human health and longevity. In this study, we present cryogenic electron microscopy structures of both ...The taurine transporter (TAUT) mediates cellular taurine uptake, playing a critical role in human health and longevity. In this study, we present cryogenic electron microscopy structures of both mouse and human TAUT in various conformational states. The taurine-bound, occluded forms of mouse and human TAUT reveal the substrate binding pocket and the ion binding sites. The amino group of taurine interacts with Glu406 at the binding site, constituting a key structural feature determining substrate preference. While both imidazole acetic acid and guanidinoethyl sulfonate (GES) inhibit TAUT by competing with taurine for the binding site, GES also functions as a substrate of TAUT. Moreover, mouse TAUT is captured in an inward-open apo conformation, where the tilted movement of transmembrane helix (TM) 1a opens the intracellular gate. Notably, TM6 exhibits two distinct conformational states: the canonical form consisting of two half-helices and a continuous straight helix. In the latter conformation, TM6 partially occupies the substrate binding site, likely promoting taurine release. Together, our findings provide critical insights into the molecular mechanisms by which TAUT recognizes and transports taurine. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_62438.map.gz emd_62438.map.gz | 203.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-62438-v30.xml emd-62438-v30.xml emd-62438.xml emd-62438.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_62438.png emd_62438.png | 43.7 KB | ||
| Filedesc metadata |  emd-62438.cif.gz emd-62438.cif.gz | 6.5 KB | ||
| Others |  emd_62438_half_map_1.map.gz emd_62438_half_map_1.map.gz emd_62438_half_map_2.map.gz emd_62438_half_map_2.map.gz | 200.2 MB 200.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-62438 http://ftp.pdbj.org/pub/emdb/structures/EMD-62438 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-62438 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-62438 | HTTPS FTP |
-Validation report
| Summary document |  emd_62438_validation.pdf.gz emd_62438_validation.pdf.gz | 801.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_62438_full_validation.pdf.gz emd_62438_full_validation.pdf.gz | 800.8 KB | Display | |
| Data in XML |  emd_62438_validation.xml.gz emd_62438_validation.xml.gz | 15.6 KB | Display | |
| Data in CIF |  emd_62438_validation.cif.gz emd_62438_validation.cif.gz | 18.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-62438 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-62438 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-62438 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-62438 | HTTPS FTP |
-Related structure data
| Related structure data |  9kmmMC  9kmiC  9kmjC  9kmkC  9kmlC  9kyoC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_62438.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_62438.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_62438_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_62438_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : TAUT
| Entire | Name: TAUT |
|---|---|
| Components |
|
-Supramolecule #1: TAUT
| Supramolecule | Name: TAUT / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: heavy chain of 9D5 fab
| Macromolecule | Name: heavy chain of 9D5 fab / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.138654 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: EVQLVESGGG LVKPGGSLKL SCAASGFTFS SYAMSWVRQS PEKRLEWVAE ISSGGRYIYY SDTVTGRFTI SRDNARNILH LEMSSLRSE DTAMYYCARG EVRQRGFDYW GQGTTLTVS |
-Macromolecule #2: Sodium- and chloride-dependent taurine transporter
| Macromolecule | Name: Sodium- and chloride-dependent taurine transporter / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 70.051891 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MATKEKLQCL KDFHKDILKP SPGKSPGTRP EDEADGKPPQ REKWSSKIDF VLSVAGGFVG LGNVWRFPYL CYKNGGGAFL IPYFIFLFG SGLPVFFLEV IIGQYTSEGG ITCWEKICPL FSGIGYASIV IVSLLNVYYI VILAWATYYL FHSFQKDLPW A HCNHSWNT ...String: MATKEKLQCL KDFHKDILKP SPGKSPGTRP EDEADGKPPQ REKWSSKIDF VLSVAGGFVG LGNVWRFPYL CYKNGGGAFL IPYFIFLFG SGLPVFFLEV IIGQYTSEGG ITCWEKICPL FSGIGYASIV IVSLLNVYYI VILAWATYYL FHSFQKDLPW A HCNHSWNT PQCMEDTLRR NESHWVSLST ANFTSPVIEF WERNVLSLSS GIDNPGSLKW DLALCLLLVW LVCFFCIWKG VR STGKVVY FTATFPFAML LVLLVRGLTL PGAGEGIKFY LYPDISRLGD PQVWIDAGTQ IFFSYAICLG AMTSLGSYNK YKY NSYRDC MLLGCLNSGT SFVSGFAIFS ILGFMAQEQG VDIADVAESG PGLAFIAYPK AVTMMPLPTF WSILFFIMLL LLGL DSQFV EVEGQITSLV DLYPSFLRKG YRREIFIAIL CSISYLLGLT MVTEGGMYVF QLFDYYAASG VCLLWVAFFE CFVIA WIYG VNRFSEDIRD MIGYRPGPWM KYSWAVITPA LCVGCFVFSL VKYVPLTYNK VYRYPDWAIG LGWGLALSSM LCIPLV IVI LLCRTEGPLR VRIKYLITPR EPNRWAVERE GATPFHSRVT LMNGALMKPS HVIVETMM UniProtKB: Sodium- and chloride-dependent taurine transporter |
-Macromolecule #3: light chain of 9D5 fab
| Macromolecule | Name: light chain of 9D5 fab / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.480681 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ENVLTQSPAI MSTSPGEKVT MTCRASSSVG SSYLHWYQQK SGASPKLWIY STSNLASGVP ARFSGSGSGT SYSLTISSVE AEDAATYYC QQFSGYPLTF GSGTKLEMK |
-Macromolecule #4: CHLORIDE ION
| Macromolecule | Name: CHLORIDE ION / type: ligand / ID: 4 / Number of copies: 1 / Formula: CL |
|---|---|
| Molecular weight | Theoretical: 35.453 Da |
-Macromolecule #5: 2H-IMIDAZOL-4-YLACETIC ACID
| Macromolecule | Name: 2H-IMIDAZOL-4-YLACETIC ACID / type: ligand / ID: 5 / Number of copies: 1 / Formula: IZC |
|---|---|
| Molecular weight | Theoretical: 126.113 Da |
| Chemical component information | 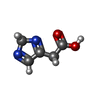 ChemComp-IZC: |
-Macromolecule #6: SODIUM ION
| Macromolecule | Name: SODIUM ION / type: ligand / ID: 6 / Number of copies: 2 |
|---|---|
| Molecular weight | Theoretical: 22.99 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 54.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.7 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)




































