+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 spike trimer (6P) in complex with three R1-26 Fabs | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Spike protein / RBD / Antibody / Fab / Viral protein / VIRAL PROTEIN-IMMUNE SYSTEM complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationMaturation of spike protein / viral translation / Translation of Structural Proteins / host cell surface / Virion Assembly and Release / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell ...Maturation of spike protein / viral translation / Translation of Structural Proteins / host cell surface / Virion Assembly and Release / host extracellular space / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / entry receptor-mediated virion attachment to host cell / membrane fusion / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / Attachment and Entry / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | ||||||||||||
| Biological species |   Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | ||||||||||||
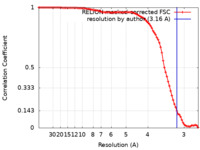
| Method | single particle reconstruction / cryo EM / Resolution: 3.16 Å | ||||||||||||
 Authors Authors | Yan Q / Gao X / Liu B / Hou R / He P / Li Z / Chen Q / Wang J / He J / Chen L ...Yan Q / Gao X / Liu B / Hou R / He P / Li Z / Chen Q / Wang J / He J / Chen L / Zhao J / Xiong X | ||||||||||||
| Funding support |  China, 3 items China, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: Antibodies utilizing VL6-57 light chains target a convergent cryptic epitope on SARS-CoV-2 spike protein and potentially drive the genesis of Omicron variants. Authors: Qihong Yan / Xijie Gao / Banghui Liu / Ruitian Hou / Ping He / Yong Ma / Yudi Zhang / Yanjun Zhang / Zimu Li / Qiuluan Chen / Jingjing Wang / Xiaohan Huang / Huan Liang / Huiran Zheng / ...Authors: Qihong Yan / Xijie Gao / Banghui Liu / Ruitian Hou / Ping He / Yong Ma / Yudi Zhang / Yanjun Zhang / Zimu Li / Qiuluan Chen / Jingjing Wang / Xiaohan Huang / Huan Liang / Huiran Zheng / Yichen Yao / Xianying Chen / Xuefeng Niu / Jun He / Ling Chen / Jincun Zhao / Xiaoli Xiong /  Abstract: Continued evolution of SARS-CoV-2 generates variants to challenge antibody immunity established by infection and vaccination. A connection between population immunity and genesis of virus variants ...Continued evolution of SARS-CoV-2 generates variants to challenge antibody immunity established by infection and vaccination. A connection between population immunity and genesis of virus variants has long been suggested but its molecular basis remains poorly understood. Here, we identify a class of SARS-CoV-2 neutralizing public antibodies defined by their shared usage of VL6-57 light chains. Although heavy chains of diverse genotypes are utilized, convergent HCDR3 rearrangements have been observed among these public antibodies to cooperate with germline VL6-57 LCDRs to target a convergent epitope defined by RBD residues S371-S373-S375. Antibody repertoire analysis identifies that this class of VL6-57 antibodies is present in SARS-CoV-2-naive individuals and is clonally expanded in most COVID-19 patients. We confirm that Omicron-specific substitutions at S371, S373 and S375 mediate escape of antibodies of the VL6-57 class. These findings support that this class of public antibodies constitutes a potential immune pressure promoting the introduction of S371L/F-S373P-S375F in Omicron variants. The results provide further molecular evidence to support that antigenic evolution of SARS-CoV-2 is driven by antibody mediated population immunity. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_60100.map.gz emd_60100.map.gz | 32.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-60100-v30.xml emd-60100-v30.xml emd-60100.xml emd-60100.xml | 21.3 KB 21.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_60100_fsc.xml emd_60100_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_60100.png emd_60100.png | 63.9 KB | ||
| Filedesc metadata |  emd-60100.cif.gz emd-60100.cif.gz | 7.2 KB | ||
| Others |  emd_60100_half_map_1.map.gz emd_60100_half_map_1.map.gz emd_60100_half_map_2.map.gz emd_60100_half_map_2.map.gz | 59.4 MB 59.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-60100 http://ftp.pdbj.org/pub/emdb/structures/EMD-60100 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60100 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-60100 | HTTPS FTP |
-Validation report
| Summary document |  emd_60100_validation.pdf.gz emd_60100_validation.pdf.gz | 787.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_60100_full_validation.pdf.gz emd_60100_full_validation.pdf.gz | 786.9 KB | Display | |
| Data in XML |  emd_60100_validation.xml.gz emd_60100_validation.xml.gz | 15.8 KB | Display | |
| Data in CIF |  emd_60100_validation.cif.gz emd_60100_validation.cif.gz | 20.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60100 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60100 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60100 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-60100 | HTTPS FTP |
-Related structure data
| Related structure data |  8zheMC  8zhdC  8zhfC  8zhgC  8zhhC  8zhiC  8zhjC  8zhkC  8zhlC  8zhmC  8zhnC  8zhoC  8zhpC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_60100.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_60100.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.3688 Å | ||||||||||||||||||||||||||||||||||||
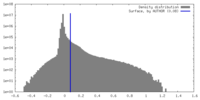
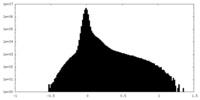
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_60100_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
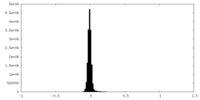
| Density Histograms |
-Half map: #2
| File | emd_60100_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
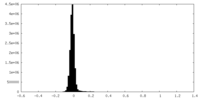
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 spike trimer (6P) in complex with three R1-26 Fabs
| Entire | Name: SARS-CoV-2 spike trimer (6P) in complex with three R1-26 Fabs |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 spike trimer (6P) in complex with three R1-26 Fabs
| Supramolecule | Name: SARS-CoV-2 spike trimer (6P) in complex with three R1-26 Fabs type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: SARS-CoV-2 spike trimer (6P)
| Supramolecule | Name: SARS-CoV-2 spike trimer (6P) / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: R1-26 Fab
| Supramolecule | Name: R1-26 Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Spike glycoprotein,Fibritin,Expression Tag
| Macromolecule | Name: Spike glycoprotein,Fibritin,Expression Tag / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 141.253875 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV YSSANNCTFE Y VSQPFLMD ...String: VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHAIHV SGTNGTKRFD NPVLPFNDGV YFASTEKSN IIRGWIFGTT LDSKTQSLLI VNNATNVVIK VCEFQFCNDP FLGVYYHKNN KSWMESEFRV YSSANNCTFE Y VSQPFLMD LEGKQGNFKN LREFVFKNID GYFKIYSKHT PINLVRDLPQ GFSALEPLVD LPIGINITRF QTLLALHRSY LT PGDSSSG WTAGAAAYYV GYLQPRTFLL KYNENGTITD AVDCALDPLS ETKCTLKSFT VEKGIYQTSN FRVQPTESIV RFP NITNLC PFGEVFNATR FASVYAWNRK RISNCVADYS VLYNSASFST FKCYGVSPTK LNDLCFTNVY ADSFVIRGDE VRQI APGQT GKIADYNYKL PDDFTGCVIA WNSNNLDSKV GGNYNYLYRL FRKSNLKPFE RDISTEIYQA GSTPCNGVEG FNCYF PLQS YGFQPTNGVG YQPYRVVVLS FELLHAPATV CGPKKSTNLV KNKCVNFNFN GLTGTGVLTE SNKKFLPFQQ FGRDIA DTT DAVRDPQTLE ILDITPCSFG GVSVITPGTN TSNQVAVLYQ DVNCTEVPVA IHADQLTPTW RVYSTGSNVF QTRAGCL IG AEHVNNSYEC DIPIGAGICA SYQTQTNSPG SASSVASQSI IAYTMSLGAE NSVAYSNNSI AIPTNFTISV TTEILPVS M TKTSVDCTMY ICGDSTECSN LLLQYGSFCT QLNRALTGIA VEQDKNTQEV FAQVKQIYKT PPIKDFGGFN FSQILPDPS KPSKRSPIED LLFNKVTLAD AGFIKQYGDC LGDIAARDLI CAQKFNGLTV LPPLLTDEMI AQYTSALLAG TITSGWTFGA GPALQIPFP MQMAYRFNGI GVTQNVLYEN QKLIANQFNS AIGKIQDSLS STPSALGKLQ DVVNQNAQAL NTLVKQLSSN F GAISSVLN DILSRLDPPE AEVQIDRLIT GRLQSLQTYV TQQLIRAAEI RASANLAATK MSECVLGQSK RVDFCGKGYH LM SFPQSAP HGVVFLHVTY VPAQEKNFTT APAICHDGKA HFPREGVFVS NGTHWFVTQR NFYEPQIITT DNTFVSGNCD VVI GIVNNT VYDPLQPELD SFKEELDKYF KNHTSPDVDL GDISGINASV VNIQKEIDRL NEVAKNLNES LIDLQELGKY EQGS GYIPE APRDGQAYVR KDGEWVLLST FLGRSLEVLF QGPGHHHHHH HHSAWSHPQF EKGGGSGGGG SGGSAWSHPQ FEK UniProtKB: Spike glycoprotein, Fibritin |
-Macromolecule #2: Heavy chain of R1-26 Fab
| Macromolecule | Name: Heavy chain of R1-26 Fab / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.908293 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGWSLILLFL VAVATRVLSE VQLVESGGGL VQPGGSLRLS CAASGFTFSS YWMSWVRQAP GKGLEWVANI KQDGSEKYYV DSVKGRFTI SRDNAKNSLY LQMNSLRAED TAVYYCARGQ LGPWVGVDYW GQGTLVTVSS ASTKGPSVFP LAPSSKSTSG G TAALGCLV ...String: MGWSLILLFL VAVATRVLSE VQLVESGGGL VQPGGSLRLS CAASGFTFSS YWMSWVRQAP GKGLEWVANI KQDGSEKYYV DSVKGRFTI SRDNAKNSLY LQMNSLRAED TAVYYCARGQ LGPWVGVDYW GQGTLVTVSS ASTKGPSVFP LAPSSKSTSG G TAALGCLV KDYFPEPVTV SWNSGALTSG VHTFPAVLQS SGLYSLSSVV TVPSSSLGTQ TYICNVNHKP SNTKVDKKVE PK SCD |
-Macromolecule #3: Light chain of R1-26 Fab
| Macromolecule | Name: Light chain of R1-26 Fab / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.558242 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MGWSCIILFL VATATGVNFM LTQPHSVSES PGKTVTISCT GSSGSIASNY VQWYQQRPGS APTTVIYEDN QRPSGVPDRF SGSIDSSSN SASLTISGLK TEDEADYYCQ SYDSSNWVFG GGTQLTVLGT KLTVLGQPKA APSVTLFPPS SEELQANKAT L VCLISDFY ...String: MGWSCIILFL VATATGVNFM LTQPHSVSES PGKTVTISCT GSSGSIASNY VQWYQQRPGS APTTVIYEDN QRPSGVPDRF SGSIDSSSN SASLTISGLK TEDEADYYCQ SYDSSNWVFG GGTQLTVLGT KLTVLGQPKA APSVTLFPPS SEELQANKAT L VCLISDFY PGAVTVAWKA DSSPVKAGVE TTTPSKQSNN KYAASSYLSL TPEQWKSHRS YSCQVTHEGS TVEKTVAPTE CS |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 30 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 295 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 81000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)