+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Ex vivo cannulae fiber structure of Pyrodictium abyssi | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Nanotube Archaea Helical protein fiber DNA-binding / PROTEIN FIBRIL | |||||||||
| Biological species |   Pyrodictium abyssi DSM 6158 (archaea) Pyrodictium abyssi DSM 6158 (archaea) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 2.3 Å | |||||||||
 Authors Authors | Sleutel M / Remaut H | |||||||||
| Funding support |  Belgium, 1 items Belgium, 1 items
| |||||||||
 Citation Citation |  Journal: bioRxiv / Year: 2024 Journal: bioRxiv / Year: 2024Title: Donor Strand Complementation and Calcium Ion Coordination Drive the Chaperone-free Polymerization of Archaeal Cannulae. Authors: Mike Sleutel / Ravi R Sonani / Jessalyn G Miller / Fengbin Wang / Andres Gonzalez Socorro / Yang Chen / Reece Martin / Borries Demeler / Michael J Rudolph / Vikram Alva / Han Remaut / Edward ...Authors: Mike Sleutel / Ravi R Sonani / Jessalyn G Miller / Fengbin Wang / Andres Gonzalez Socorro / Yang Chen / Reece Martin / Borries Demeler / Michael J Rudolph / Vikram Alva / Han Remaut / Edward H Egelman / Vincent P Conticello /     Abstract: Cannulae are tubular protein filaments that accumulate on the extracellular surface of the hyperthermophilic archaeon during cell division. Cannulae have been postulated to act as a primitive ...Cannulae are tubular protein filaments that accumulate on the extracellular surface of the hyperthermophilic archaeon during cell division. Cannulae have been postulated to act as a primitive extracellular matrix through which cells could communicate or exchange material, although their native biological function remains obscure. Here, we report cryoEM structural analyses of cannulae and of protein assemblies derived from recombinant cannula-like proteins. Three-dimensional reconstructions of cannulae revealed that the structural interactions between protomers in the native and recombinant filaments were based on donor strand complementation, a form of non-covalent polymerization in which a donor β-strand from one subunit is inserted into an acceptor groove in a β-sheet of a neighboring subunit. Donor strand complementation in cannulae is reinforced through calcium ion coordination at the interfaces between structural subunits in the respective assemblies. While donor strand complementation occurs during the assembly of chaperone-usher pili, this process requires the participation of accessory proteins that are localized in the outer membrane. In contrast, we demonstrate that calcium ions can induce assembly of cannulae in the absence of other co-factors. Crystallographic analysis of a recombinant cannula-like protein monomer provided evidence that calcium ion binding primes the precursor for donor strand invasion through unblocking of the acceptor groove. Bioinformatic analysis suggested that structurally homologous cannula-like proteins occurred within the genomes of other hyperthermophilic archaea and were encompassed within the TasA superfamily of biomatrix proteins. CryoEM structural analyses of tubular filaments derived from assembly of a recombinant cannula-like protein from an uncultured species revealed a common mode of assembly to the cannulae, in which donor strand complementation and calcium ion binding stabilized longitudinal and lateral assembly in tubular 2D sheets. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_51935.map.gz emd_51935.map.gz | 236.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-51935-v30.xml emd-51935-v30.xml emd-51935.xml emd-51935.xml | 14.6 KB 14.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_51935_fsc.xml emd_51935_fsc.xml | 19.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_51935.png emd_51935.png | 216.8 KB | ||
| Masks |  emd_51935_msk_1.map emd_51935_msk_1.map | 824 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-51935.cif.gz emd-51935.cif.gz | 5.9 KB | ||
| Others |  emd_51935_half_map_1.map.gz emd_51935_half_map_1.map.gz emd_51935_half_map_2.map.gz emd_51935_half_map_2.map.gz | 764.6 MB 764.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-51935 http://ftp.pdbj.org/pub/emdb/structures/EMD-51935 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-51935 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-51935 | HTTPS FTP |
-Validation report
| Summary document |  emd_51935_validation.pdf.gz emd_51935_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_51935_full_validation.pdf.gz emd_51935_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_51935_validation.xml.gz emd_51935_validation.xml.gz | 29.5 KB | Display | |
| Data in CIF |  emd_51935_validation.cif.gz emd_51935_validation.cif.gz | 38.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-51935 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-51935 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-51935 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-51935 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_51935.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_51935.map.gz / Format: CCP4 / Size: 824 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.71 Å | ||||||||||||||||||||||||||||||||||||
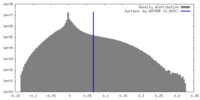
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_51935_msk_1.map emd_51935_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
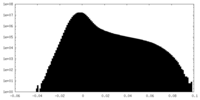
| Density Histograms |
-Half map: #1
| File | emd_51935_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_51935_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Ex vivo CanX fibers from Pyrodictium abyssi
| Entire | Name: Ex vivo CanX fibers from Pyrodictium abyssi |
|---|---|
| Components |
|
-Supramolecule #1: Ex vivo CanX fibers from Pyrodictium abyssi
| Supramolecule | Name: Ex vivo CanX fibers from Pyrodictium abyssi / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:   Pyrodictium abyssi DSM 6158 (archaea) Pyrodictium abyssi DSM 6158 (archaea) |
-Macromolecule #1: CanX
| Macromolecule | Name: CanX / type: protein_or_peptide / ID: 1 / Number of copies: 154 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Pyrodictium abyssi DSM 6158 (archaea) Pyrodictium abyssi DSM 6158 (archaea) |
| Molecular weight | Theoretical: 18.745406 KDa |
| Recombinant expression | Organism:   Pyrodictium abyssi DSM 6158 (archaea) Pyrodictium abyssi DSM 6158 (archaea) |
| Sequence | String: MRYTTLAIAG IIASAAALAL LAGFATTQSP LNSFYATGTA QAVQEPIDVE SHLDNTIAPA AGAQGYKDMG YVKIINYTDV NVVKLKVTL ANAAQLRPYF KYLQLVLTSN ASSTVEETKA VLSLKKPSAV IILDNDDYSS TNKIQLKVEA YYEAKEGMLF D SLPVILNF QVLSVS |
-Macromolecule #2: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 2 / Number of copies: 462 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 3 / Number of copies: 154 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 5.5 |
|---|---|
| Grid | Model: Quantifoil R2/1 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | JEOL CRYO ARM 300 |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.6 µm / Nominal defocus min: 0.8 µm |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)