[English] 日本語
 Yorodumi
Yorodumi- EMDB-5168: Direct visualization of secondary structures of F-actin by electr... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-5168 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Direct visualization of secondary structures of F-actin by electron cryomicroscopy | |||||||||
 Map data Map data | CryoEM map of F-actin at 6.6 angstrom resolution | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | F-actin | |||||||||
| Function / homology |  Function and homology information Function and homology informationcytoskeletal motor activator activity / tropomyosin binding / mesenchyme migration / troponin I binding / myosin heavy chain binding / actin filament bundle / filamentous actin / skeletal muscle thin filament assembly / striated muscle thin filament / actin filament bundle assembly ...cytoskeletal motor activator activity / tropomyosin binding / mesenchyme migration / troponin I binding / myosin heavy chain binding / actin filament bundle / filamentous actin / skeletal muscle thin filament assembly / striated muscle thin filament / actin filament bundle assembly / skeletal muscle myofibril / actin monomer binding / skeletal muscle fiber development / stress fiber / titin binding / actin filament polymerization / filopodium / actin filament / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / calcium-dependent protein binding / lamellipodium / cell body / hydrolase activity / protein domain specific binding / calcium ion binding / positive regulation of gene expression / magnesium ion binding / ATP binding / identical protein binding / cytoplasm Similarity search - Function | |||||||||
| Biological species | unidentified (others) | |||||||||
| Method | helical reconstruction / cryo EM / Resolution: 6.6 Å | |||||||||
 Authors Authors | Fujii T / Iwane AH / Yanagida T / Namba K | |||||||||
 Citation Citation |  Journal: Nature / Year: 2010 Journal: Nature / Year: 2010Title: Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Authors: Takashi Fujii / Atsuko H Iwane / Toshio Yanagida / Keiichi Namba /  Abstract: F-actin is a helical assembly of actin, which is a component of muscle fibres essential for contraction and has a crucial role in numerous cellular processes, such as the formation of lamellipodia ...F-actin is a helical assembly of actin, which is a component of muscle fibres essential for contraction and has a crucial role in numerous cellular processes, such as the formation of lamellipodia and filopodia, as the most abundant component and regulator of cytoskeletons by dynamic assembly and disassembly (from G-actin to F-actin and vice versa). Actin is a ubiquitous protein and is involved in important biological functions, but the definitive high-resolution structure of F-actin remains unknown. Although a recent atomic model well reproduced X-ray fibre diffraction intensity data from a highly oriented liquid-crystalline sol specimen, its refinement without experimental phase information has certain limitations. Direct visualization of the structure by electron cryomicroscopy, however, has been difficult because it is relatively thin and flexible. Here we report the F-actin structure at 6.6 Å resolution, made obtainable by recent advances in electron cryomicroscopy. The density map clearly resolves all the secondary structures of G-actin, such as α-helices, β-structures and loops, and makes unambiguous modelling and refinement possible. Complex domain motions that open the nucleotide-binding pocket on F-actin formation, specific D-loop and terminal conformations, and relatively tight axial but markedly loose interprotofilament interactions hydrophilic in nature are revealed in the F-actin model, and all seem to be important for dynamic functions of actin. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_5168.map.gz emd_5168.map.gz | 3.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-5168-v30.xml emd-5168-v30.xml emd-5168.xml emd-5168.xml | 9.4 KB 9.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_5168_1.jpg emd_5168_1.jpg | 47.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-5168 http://ftp.pdbj.org/pub/emdb/structures/EMD-5168 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5168 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-5168 | HTTPS FTP |
-Validation report
| Summary document |  emd_5168_validation.pdf.gz emd_5168_validation.pdf.gz | 344.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_5168_full_validation.pdf.gz emd_5168_full_validation.pdf.gz | 344.4 KB | Display | |
| Data in XML |  emd_5168_validation.xml.gz emd_5168_validation.xml.gz | 5.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5168 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5168 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5168 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-5168 | HTTPS FTP |
-Related structure data
| Related structure data |  3mfpMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_5168.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_5168.map.gz / Format: CCP4 / Size: 3.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM map of F-actin at 6.6 angstrom resolution | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.742 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
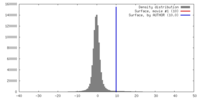
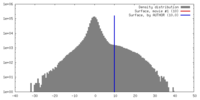
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : F-actin
| Entire | Name: F-actin |
|---|---|
| Components |
|
-Supramolecule #1000: F-actin
| Supramolecule | Name: F-actin / type: sample / ID: 1000 / Oligomeric state: helical structure / Number unique components: 7 |
|---|
-Macromolecule #1: F-actin
| Macromolecule | Name: F-actin / type: protein_or_peptide / ID: 1 / Name.synonym: F-actin / Recombinant expression: No / Database: NCBI |
|---|---|
| Source (natural) | Organism: unidentified (others) / synonym: rabbit |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 Details: 25 mM Hepes buffer (pH 7.5), 50 mM KCl, 1 mM MgCl2 and 1 mM ATP |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Instrument: OTHER / Details: Vitrification instrument: Vitrobot / Method: Blot for 3.5 seconds before plunging |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 3200FSC |
|---|---|
| Temperature | Average: 50 K |
| Specialist optics | Energy filter - Name: Omega filter / Energy filter - Lower energy threshold: 0.0 eV / Energy filter - Upper energy threshold: 10.0 eV |
| Date | Mar 21, 2009 |
| Image recording | Category: CCD / Film or detector model: TVIPS TEMCAM-F415 (4k x 4k) / Digitization - Sampling interval: 15 µm / Number real images: 490 / Average electron dose: 20 e/Å2 / Bits/pixel: 16 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 172000 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 1.6 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 100000 |
| Sample stage | Specimen holder: Top entry liquid helium-cooled cryo specimen holder Specimen holder model: JEOL 3200FSC CRYOHOLDER |
- Image processing
Image processing
| Details | 1-start helix (166.6 degree, 27.6 angstrom) is left handed. F-actin looks like a two strand ribbon. The long pitch helix along the strand of F-actin is right handed. |
|---|---|
| Final reconstruction | Applied symmetry - Helical parameters - Δz: 27.6 Å Applied symmetry - Helical parameters - Δ&Phi: 166.6 ° Resolution.type: BY AUTHOR / Resolution: 6.6 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: SPIDER |
| CTF correction | Details: Each particle |
 Movie
Movie Controller
Controller














 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)