[English] 日本語
 Yorodumi
Yorodumi- EMDB-50169: Cryo-EM structure of Dopamine 3 receptor:Go complex bound to bito... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of Dopamine 3 receptor:Go complex bound to bitopic FOB02-04A - Conformation B | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / complex / bitopic / membrane protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationmusculoskeletal movement, spinal reflex action / acid secretion / dopamine neurotransmitter receptor activity, coupled via Gi/Go / response to histamine / regulation of potassium ion transport / adenylate cyclase-inhibiting dopamine receptor signaling pathway / Dopamine receptors / regulation of dopamine uptake involved in synaptic transmission / positive regulation of dopamine receptor signaling pathway / phospholipase C-activating dopamine receptor signaling pathway ...musculoskeletal movement, spinal reflex action / acid secretion / dopamine neurotransmitter receptor activity, coupled via Gi/Go / response to histamine / regulation of potassium ion transport / adenylate cyclase-inhibiting dopamine receptor signaling pathway / Dopamine receptors / regulation of dopamine uptake involved in synaptic transmission / positive regulation of dopamine receptor signaling pathway / phospholipase C-activating dopamine receptor signaling pathway / negative regulation of oligodendrocyte differentiation / mu-type opioid receptor binding / corticotropin-releasing hormone receptor 1 binding / negative regulation of synaptic transmission, glutamatergic / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Ca2+ pathway / G protein-coupled receptor internalization / G alpha (z) signalling events / Vasopressin regulates renal water homeostasis via Aquaporins / vesicle docking involved in exocytosis / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / G alpha (q) signalling events / Thrombin signalling through proteinase activated receptors (PARs) / G alpha (i) signalling events / alkylglycerophosphoethanolamine phosphodiesterase activity / response to morphine / photoreceptor outer segment membrane / arachidonate secretion / G protein-coupled dopamine receptor signaling pathway / regulation of heart contraction / spectrin binding / positive regulation of cytokinesis / negative regulation of cytosolic calcium ion concentration / regulation of dopamine secretion / dopamine metabolic process / behavioral response to cocaine / social behavior / negative regulation of protein secretion / negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / photoreceptor outer segment / negative regulation of insulin secretion / prepulse inhibition / G protein-coupled serotonin receptor binding / adenylate cyclase-activating adrenergic receptor signaling pathway / cardiac muscle cell apoptotic process / muscle contraction / negative regulation of blood pressure / photoreceptor inner segment / positive regulation of mitotic nuclear division / bioluminescence / learning / response to cocaine / generation of precursor metabolites and energy / locomotory behavior / G protein-coupled receptor activity / circadian regulation of gene expression / visual learning / G-protein beta/gamma-subunit complex binding / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / intracellular calcium ion homeostasis / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / sensory perception of taste / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / cellular response to prostaglandin E stimulus / G-protein beta-subunit binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / signaling receptor complex adaptor activity Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.09 Å | |||||||||
 Authors Authors | Arroyo-Urea S / Garcia-Nafria J | |||||||||
| Funding support |  Spain, 1 items Spain, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2024 Journal: Nat Commun / Year: 2024Title: A bitopic agonist bound to the dopamine 3 receptor reveals a selectivity site. Authors: Sandra Arroyo-Urea / Antonina L Nazarova / Ángela Carrión-Antolí / Alessandro Bonifazi / Francisco O Battiti / Jordy Homing Lam / Amy Hauck Newman / Vsevolod Katritch / Javier García-Nafría /   Abstract: Although aminergic GPCRs are the target for ~25% of approved drugs, developing subtype selective drugs is a major challenge due to the high sequence conservation at their orthosteric binding site. ...Although aminergic GPCRs are the target for ~25% of approved drugs, developing subtype selective drugs is a major challenge due to the high sequence conservation at their orthosteric binding site. Bitopic ligands are covalently joined orthosteric and allosteric pharmacophores with the potential to boost receptor selectivity and improve current medications by reducing off-target side effects. However, the lack of structural information on their binding mode impedes rational design. Here we determine the cryo-EM structure of the hDR:Gαβγ complex bound to the DR selective bitopic agonist FOB02-04A. Structural, functional and computational analyses provide insights into its binding mode and point to a new TM2-ECL1-TM1 region, which requires the N-terminal ordering of TM1, as a major determinant of subtype selectivity in aminergic GPCRs. This region is underexploited in drug development, expands the established secondary binding pocket in aminergic GPCRs and could potentially be used to design novel and subtype selective drugs. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_50169.map.gz emd_50169.map.gz | 95.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-50169-v30.xml emd-50169-v30.xml emd-50169.xml emd-50169.xml | 23.5 KB 23.5 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_50169.png emd_50169.png | 34.4 KB | ||
| Masks |  emd_50169_msk_1.map emd_50169_msk_1.map emd_50169_msk_2.map emd_50169_msk_2.map | 103 MB 103 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-50169.cif.gz emd-50169.cif.gz | 7.6 KB | ||
| Others |  emd_50169_half_map_1.map.gz emd_50169_half_map_1.map.gz emd_50169_half_map_2.map.gz emd_50169_half_map_2.map.gz | 95.7 MB 95.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-50169 http://ftp.pdbj.org/pub/emdb/structures/EMD-50169 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50169 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-50169 | HTTPS FTP |
-Validation report
| Summary document |  emd_50169_validation.pdf.gz emd_50169_validation.pdf.gz | 844.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_50169_full_validation.pdf.gz emd_50169_full_validation.pdf.gz | 844.5 KB | Display | |
| Data in XML |  emd_50169_validation.xml.gz emd_50169_validation.xml.gz | 13.2 KB | Display | |
| Data in CIF |  emd_50169_validation.cif.gz emd_50169_validation.cif.gz | 15.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50169 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50169 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50169 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-50169 | HTTPS FTP |
-Related structure data
| Related structure data |  9f34MC  9f33C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_50169.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_50169.map.gz / Format: CCP4 / Size: 103 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.84 Å | ||||||||||||||||||||||||||||||||||||
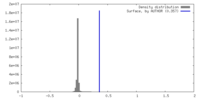
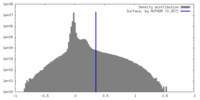
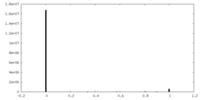
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_50169_msk_1.map emd_50169_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
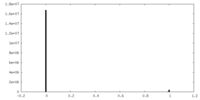
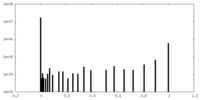
| Density Histograms |
-Mask #2
| File |  emd_50169_msk_2.map emd_50169_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
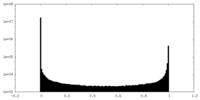
| Density Histograms |
- Sample components
Sample components
-Entire : D3R: Gtrimer: scFv16 complex bound to the bitopic FOB02-04A - Con...
| Entire | Name: D3R: Gtrimer: scFv16 complex bound to the bitopic FOB02-04A - Conformation B |
|---|---|
| Components |
|
-Supramolecule #1: D3R: Gtrimer: scFv16 complex bound to the bitopic FOB02-04A - Con...
| Supramolecule | Name: D3R: Gtrimer: scFv16 complex bound to the bitopic FOB02-04A - Conformation B type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 190 KDa |
-Macromolecule #1: Guanine nucleotide-binding protein G(o) subunit alpha
| Macromolecule | Name: Guanine nucleotide-binding protein G(o) subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.0975 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MGCTLSAEER AALERSKAIE KNLKEDGISA AKDVKLLLLG AGESGKNTIV KQMKIIHEDG FSGEDVKQYK PVVYSNTIQS LAAIVRAMD TLGIEYGDKE RKADAKMVCD VVSRMEDTEP FSAELLSAMM RLWGDSGIQE CFNRSREYQL NDSAKYYLDS L DRIGAADY ...String: MGCTLSAEER AALERSKAIE KNLKEDGISA AKDVKLLLLG AGESGKNTIV KQMKIIHEDG FSGEDVKQYK PVVYSNTIQS LAAIVRAMD TLGIEYGDKE RKADAKMVCD VVSRMEDTEP FSAELLSAMM RLWGDSGIQE CFNRSREYQL NDSAKYYLDS L DRIGAADY QPTEQDILRT RVKTTGIVET HFTFKNLHFR LFDVGAQRSE RKKWIHCFED VTAIIFCVAL SGYDQVLHED ET TNRMHAS LKLFDSICNN KFFIDTSIIL FLNKKDLFGE KIKKSPLTIC FPEYTGPNTY EDAAAYIQAQ FESKNRSPNK EIY CHMTCS TDTNNIQVVF DAVTDIIIAN NLRGCGLY UniProtKB: Guanine nucleotide-binding protein G(o) subunit alpha, Guanine nucleotide-binding protein G(o) subunit alpha |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.373992 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MHHHHHHHHE NLYFQGSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSAS QDGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG Y LSCCRFLD ...String: MHHHHHHHHE NLYFQGSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSAS QDGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG Y LSCCRFLD DNQIVTSSGD TTCALWDIET GQQTTTFTGH TGDVMSLSLA PDTRLFVSGA CDASAKLWDV REGMCRQTFT GH ESDINAI CFFPNGNAFA TGSDDATCRL FDLRADQELM TYSHDNIICG ITSVSFSKSG RLLLAGYDDF NCNVWDALKA DRA GVLAGH DNRVSCLGVT DDGMAVATGS WDSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.845078 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFSAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Antibody scFv16
| Macromolecule | Name: Antibody scFv16 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 27.625844 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MDVQLVESGG GLVQPGGSRK LSCSASGFAF SSFGMHWVRQ APEKGLEWVA YISSGSGTIY YADTVKGRFT ISRDDPKNTL FLQMTSLRS EDTAMYYCVR SIYYYGSSPF DFWGQGTTLT VSSGGGGSGG GGSGGGGSDI VMTQATSSVP VTPGESVSIS C RSSKSLLH ...String: MDVQLVESGG GLVQPGGSRK LSCSASGFAF SSFGMHWVRQ APEKGLEWVA YISSGSGTIY YADTVKGRFT ISRDDPKNTL FLQMTSLRS EDTAMYYCVR SIYYYGSSPF DFWGQGTTLT VSSGGGGSGG GGSGGGGSDI VMTQATSSVP VTPGESVSIS C RSSKSLLH SNGNTYLYWF LQRPGQSPQL LIYRMSNLAS GVPDRFSGSG SGTAFTLTIS RLEAEDVGVY YCMQHLEYPL TF GAGTKLE LKGSLEVLFQ GP |
-Macromolecule #5: Green fluorescent protein,D(3) dopamine receptor
| Macromolecule | Name: Green fluorescent protein,D(3) dopamine receptor / type: protein_or_peptide / ID: 5 Details: HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP- ...Details: HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R,HASS-FLAG-eGFP-D3R Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 74.733945 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDKVSKGEE LFTGVVPILV ELDGDVNGHK FSVSGEGEGD ATYGKLTLKF ICTTGKLPVP WPTLVTTLT YGVQCFSRYP DHMKQHDFFK SAMPEGYVQE RTIFFKDDGN YKTRAEVKFE GDTLVNRIEL KGIDFKEDGN I LGHKLEYN ...String: MKTIIALSYI FCLVFADYKD DDDKVSKGEE LFTGVVPILV ELDGDVNGHK FSVSGEGEGD ATYGKLTLKF ICTTGKLPVP WPTLVTTLT YGVQCFSRYP DHMKQHDFFK SAMPEGYVQE RTIFFKDDGN YKTRAEVKFE GDTLVNRIEL KGIDFKEDGN I LGHKLEYN YNSHNVYIMA DKQKNGIKVN FKIRHNIEDG SVQLADHYQQ NTPIGDGPVL LPDNHYLSTQ SALSKDPNEK RD HMVLLEF VTAAGITLGM DELYKLEVLF QGPASLSQLS SHLNYTCGAE NSTGASQARP HAYYALSYCA LILAIVFGNG LVC MAVLKE RALQTTTNYL VVSLAVADLL VATLVMPWVV YLEVTGGVWN FSRICCDVFV TLDVMMCTAS IWNLCAISID RYTA VVMPV HYQHGTGQSS CRRVALMITA VWVLAFAVSC PLLFGFNTTG DPTVCSISNP DFVIYSSVVS FYLPFGVTVL VYARI YVVL KQRRRKRILT RQNSQCNSVR PGFPQQTLSP DPAHLELKRY YSICQDTALG GPGFQERGGE LKREEKTRNS LSPTIA PKL SLEVRKLSNG RLSTSLKLGP LQPRGVPLRE KKATQMVAIV LGAFIVCWLP FFLTHVLNTH CQTCHVSPEL YSATTWL GY VNSALNPVIY TTFNIEFRKA FLKILSC UniProtKB: Green fluorescent protein, D(3) dopamine receptor, D(3) dopamine receptor, D(3) dopamine receptor |
-Macromolecule #6: N-[2-[(1R,2S)-2-[[(2S,5S)-2-(6-azanylpyridin-3-yl)-5-methyl-morph...
| Macromolecule | Name: N-[2-[(1R,2S)-2-[[(2S,5S)-2-(6-azanylpyridin-3-yl)-5-methyl-morpholin-4-yl]methyl]cyclopropyl]ethyl]-1H-indole-2-carboxamide type: ligand / ID: 6 / Number of copies: 1 / Formula: A1H9N |
|---|---|
| Molecular weight | Theoretical: 433.546 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.8 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R0.6/1 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: SUPER-RESOLUTION / Number real images: 22655 / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.0 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: OTHER |
|---|---|
| Output model |  PDB-9f34: |
 Movie
Movie Controller
Controller


























 Z (Sec.)
Z (Sec.) X (Row.)
X (Row.) Y (Col.)
Y (Col.)