[English] 日本語
 Yorodumi
Yorodumi- EMDB-4906: Cryo-EM reconstruction of TssA protein from T6SS of Escherichia coli. -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4906 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM reconstruction of TssA protein from T6SS of Escherichia coli. | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
| Function / homology | Type VI secretion system-associated, VCA0119 / Type VI secretion, EvfE, EvfF, ImpA, BimE, VC_A0119, VasJ / ImpA, N-terminal / ImpA, N-terminal, type VI secretion system / Putative type VI secretion protein Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
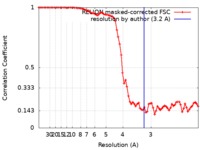
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Nazarov S / Shneider M / Basler M / Leiman P | |||||||||
| Funding support |  Switzerland, 1 items Switzerland, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-EM structure of TssA protein from Type VI secretion system of E. coli. Authors: Nazarov S / Demurtas D / Shneider M / Basler M / Leiman P | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
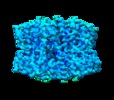
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4906.map.gz emd_4906.map.gz | 100.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4906-v30.xml emd-4906-v30.xml emd-4906.xml emd-4906.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4906_fsc.xml emd_4906_fsc.xml | 7.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_4906.png emd_4906.png | 144 KB | ||
| Masks |  emd_4906_msk_1.map emd_4906_msk_1.map | 129.7 MB |  Mask map Mask map | |
| Others |  emd_4906_additional_1.map.gz emd_4906_additional_1.map.gz emd_4906_additional_2.map.gz emd_4906_additional_2.map.gz emd_4906_half_map_1.map.gz emd_4906_half_map_1.map.gz emd_4906_half_map_2.map.gz emd_4906_half_map_2.map.gz | 37.8 MB 120.5 MB 95.6 MB 97.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4906 http://ftp.pdbj.org/pub/emdb/structures/EMD-4906 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4906 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4906 | HTTPS FTP |
-Validation report
| Summary document |  emd_4906_validation.pdf.gz emd_4906_validation.pdf.gz | 401.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_4906_full_validation.pdf.gz emd_4906_full_validation.pdf.gz | 400.2 KB | Display | |
| Data in XML |  emd_4906_validation.xml.gz emd_4906_validation.xml.gz | 16 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4906 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4906 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4906 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4906 | HTTPS FTP |
-Related structure data
| Related structure data |  6rjuMUC M: atomic model generated by this map U: unfit; in different coordinate system*YM C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4906.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4906.map.gz / Format: CCP4 / Size: 129.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.058 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Mask #1
| File |  emd_4906_msk_1.map emd_4906_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_4906_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #2
| File | emd_4906_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_4906_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_4906_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : C-terminal domain of T6SS protein TssA.
| Entire | Name: C-terminal domain of T6SS protein TssA. |
|---|---|
| Components |
|
-Supramolecule #1: C-terminal domain of T6SS protein TssA.
| Supramolecule | Name: C-terminal domain of T6SS protein TssA. / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  |
| Molecular weight | Theoretical: 634 KDa |
-Macromolecule #1: Putative type VI secretion protein
| Macromolecule | Name: Putative type VI secretion protein / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 59.159789 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASIHSLLSA CQTTPRDVAE PAQVRIALWD KWLAPVTPDN PAGDDAGYDD DFQQMREEVN KLSGADAGIV SQLAEKLLTT RTKDIRVAT WYIWARLRQD GEKGLADGLE LLTGLLQRFG EHLHPQRSRA RKAALEWLCS ARILDSLSLY PEVVKADTLR I AGALWLAE ...String: MASIHSLLSA CQTTPRDVAE PAQVRIALWD KWLAPVTPDN PAGDDAGYDD DFQQMREEVN KLSGADAGIV SQLAEKLLTT RTKDIRVAT WYIWARLRQD GEKGLADGLE LLTGLLQRFG EHLHPQRSRA RKAALEWLCS ARILDSLSLY PEVVKADTLR I AGALWLAE QTFTDEASAP VLNGLYQALE NRLMKAGGVD AVVPQEAAAP APTVTSGSVM ALSAITSGQE LLSQARVLAK YL RDQPEGW LAAHRLMKSV RHDTLHQLPP LSADGRTRIA PPGPDRRASL KRLYLQQNWL SLLEQCDDMF ARGASHLWLD LQW YIHQAL LQTGKENYAA IIQYDLKGLL LRLPGLETLA FNDGMPFADD VTLSWIQQQV MECGERWAEE PSVTITAAPG DNDI LSLEP EALQIADNEG TEAALSWLQA RPGIQSDRSN WLLRLL(MSE)ARV AEQTGKNDLA LHLLAELDER ATRLTLSQWE P ELVFEVKA RRLKLLR(MSE)KS AKTESDRVRL QPD(MSE)EHLLAG LIAIDAARAA VLCNSGSS |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 90 % / Chamber temperature: 293 K / Details: Leica EM GP2. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Number real images: 3269 / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z
Z Y
Y X
X