[English] 日本語
 Yorodumi
Yorodumi- EMDB-47628: Cryo-EM structure of the Pyrobaculum calidifontis 70S ribosome (C... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Pyrobaculum calidifontis 70S ribosome (Composite Map) | |||||||||
 Map data Map data | Pyrobaculum calidifontis 70S ribosome (composite map) | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Ribosome / Translation / Hibernation / RNA | |||||||||
| Function / homology |  Function and homology information Function and homology informationribonuclease P activity / tRNA 5'-leader removal / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / ribosomal large subunit biogenesis / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / ribosome biogenesis / large ribosomal subunit / ribosomal small subunit biogenesis / ribosomal small subunit assembly / small ribosomal subunit ...ribonuclease P activity / tRNA 5'-leader removal / maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / ribosomal large subunit biogenesis / maturation of SSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA) / ribosome biogenesis / large ribosomal subunit / ribosomal small subunit biogenesis / ribosomal small subunit assembly / small ribosomal subunit / small ribosomal subunit rRNA binding / 5S rRNA binding / ribosomal large subunit assembly / cytosolic small ribosomal subunit / large ribosomal subunit rRNA binding / cytosolic large ribosomal subunit / cytoplasmic translation / tRNA binding / negative regulation of translation / rRNA binding / ribosome / structural constituent of ribosome / translation / ribonucleoprotein complex / mRNA binding / RNA binding / zinc ion binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |   Pyrobaculum calidifontis JCM 11548 (archaea) Pyrobaculum calidifontis JCM 11548 (archaea) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.36 Å | |||||||||
 Authors Authors | Nissley AJ / Cate JHD | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Structure of an Archaeal Ribosome with a Divergent Active Site Authors: Nissley AJ / Shulgina Y / Kivimae RW / Downing BE / Penev PI / Banfield JF / Nayak DD / Cate JHD | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_47628.map.gz emd_47628.map.gz | 744.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-47628-v30.xml emd-47628-v30.xml emd-47628.xml emd-47628.xml | 95.7 KB 95.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_47628.png emd_47628.png | 154.6 KB | ||
| Filedesc metadata |  emd-47628.cif.gz emd-47628.cif.gz | 16.7 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-47628 http://ftp.pdbj.org/pub/emdb/structures/EMD-47628 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-47628 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-47628 | HTTPS FTP |
-Validation report
| Summary document |  emd_47628_validation.pdf.gz emd_47628_validation.pdf.gz | 618.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_47628_full_validation.pdf.gz emd_47628_full_validation.pdf.gz | 617.7 KB | Display | |
| Data in XML |  emd_47628_validation.xml.gz emd_47628_validation.xml.gz | 8.5 KB | Display | |
| Data in CIF |  emd_47628_validation.cif.gz emd_47628_validation.cif.gz | 9.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-47628 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-47628 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-47628 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-47628 | HTTPS FTP |
-Related structure data
| Related structure data |  9e71MC  9e6qC  9e7fC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_47628.map.gz / Format: CCP4 / Size: 857.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_47628.map.gz / Format: CCP4 / Size: 857.4 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Pyrobaculum calidifontis 70S ribosome (composite map) | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.8293 Å | ||||||||||||||||||||||||||||||||||||
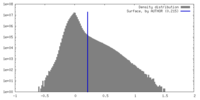
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
- Sample components
Sample components
+Entire : 70S ribosome
+Supramolecule #1: 70S ribosome
+Supramolecule #2: 50S Subunit
+Supramolecule #3: 30S subunit
+Macromolecule #1: 5S rRNA
+Macromolecule #2: 23S rRNA
+Macromolecule #3: 16S rRNA
+Macromolecule #4: Large ribosomal subunit protein uL2
+Macromolecule #5: Large ribosomal subunit protein uL3
+Macromolecule #6: Large ribosomal subunit protein uL4
+Macromolecule #7: Large ribosomal subunit protein uL5
+Macromolecule #8: Large ribosomal subunit protein uL6
+Macromolecule #9: Large ribosomal subunit protein eL8
+Macromolecule #10: Large ribosomal subunit protein uL13
+Macromolecule #11: Large ribosomal subunit protein eL13
+Macromolecule #12: Large ribosomal subunit protein uL14
+Macromolecule #13: Large ribosomal subunit protein eL14
+Macromolecule #14: Large ribosomal subunit protein uL15
+Macromolecule #15: 50S ribosomal protein L15e
+Macromolecule #16: Large ribosomal subunit protein uL16
+Macromolecule #17: Large ribosomal subunit protein uL18
+Macromolecule #18: Large ribosomal subunit protein eL18
+Macromolecule #19: Large ribosomal subunit protein eL19
+Macromolecule #20: Large ribosomal subunit protein eL20
+Macromolecule #21: Large ribosomal subunit protein eL21
+Macromolecule #22: Large ribosomal subunit protein uL22
+Macromolecule #23: Large ribosomal subunit protein uL23
+Macromolecule #24: Large ribosomal subunit protein uL24
+Macromolecule #25: Large ribosomal subunit protein eL24
+Macromolecule #26: Large ribosomal subunit protein uL29
+Macromolecule #27: Large ribosomal subunit protein uL30
+Macromolecule #28: Large ribosomal subunit protein eL30
+Macromolecule #29: Large ribosomal subunit protein eL31
+Macromolecule #30: Large ribosomal subunit protein eL32
+Macromolecule #31: Large ribosomal subunit protein eL34
+Macromolecule #32: Large ribosomal subunit protein eL37
+Macromolecule #33: LSU ribosomal protein L38E
+Macromolecule #34: Large ribosomal subunit protein eL39
+Macromolecule #35: Large ribosomal subunit protein eL40
+Macromolecule #36: eL42
+Macromolecule #37: Large ribosomal subunit protein eL43
+Macromolecule #38: DJ-1/PfpI domain-containing protein
+Macromolecule #39: PaREP1 domain containing protein
+Macromolecule #40: Small ribosomal subunit protein eS1
+Macromolecule #41: Small ribosomal subunit protein uS2
+Macromolecule #42: Small ribosomal subunit protein uS3
+Macromolecule #43: Small ribosomal subunit protein uS4
+Macromolecule #44: Small ribosomal subunit protein eS4
+Macromolecule #45: Small ribosomal subunit protein uS5
+Macromolecule #46: Small ribosomal subunit protein eS6
+Macromolecule #47: Small ribosomal subunit protein uS7
+Macromolecule #48: Small ribosomal subunit protein uS8
+Macromolecule #49: Small ribosomal subunit protein eS8
+Macromolecule #50: Small ribosomal subunit protein uS9
+Macromolecule #51: Small ribosomal subunit protein uS10
+Macromolecule #52: Small ribosomal subunit protein uS11
+Macromolecule #53: Small ribosomal subunit protein uS12
+Macromolecule #54: Small ribosomal subunit protein uS13
+Macromolecule #55: Small ribosomal subunit protein uS14
+Macromolecule #56: Small ribosomal subunit protein uS15
+Macromolecule #57: Small ribosomal subunit protein uS17
+Macromolecule #58: Small ribosomal subunit protein eS17
+Macromolecule #59: Small ribosomal subunit protein uS19
+Macromolecule #60: Small ribosomal subunit protein eS19
+Macromolecule #61: Small ribosomal subunit protein eS24
+Macromolecule #62: SSU ribosomal protein S25E
+Macromolecule #63: SSU ribosomal protein S26E
+Macromolecule #64: Small ribosomal subunit protein eS27
+Macromolecule #65: eS28
+Macromolecule #66: SSU ribosomal protein S30E
+Macromolecule #67: aS35
+Macromolecule #68: Small zinc finger protein HVO-2753-like zinc-binding pocket domai...
+Macromolecule #69: SPERMINE
+Macromolecule #70: MAGNESIUM ION
+Macromolecule #71: ZINC ION
+Macromolecule #72: water
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.12 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7 Component:
| ||||||||||||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Support film - Material: CARBON / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 7318 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 105000 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-9e71: |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)