+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Citrobacter multi-ubiquitin protein filament | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | filament / beta-grasp / PROTEIN BINDING | |||||||||
| Biological species |  Citrobacter sp. RHBSTW-00271 (bacteria) Citrobacter sp. RHBSTW-00271 (bacteria) | |||||||||
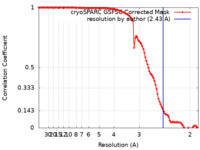
| Method | helical reconstruction / cryo EM / Resolution: 2.43 Å | |||||||||
 Authors Authors | Gong M / Gu Y / Corbett KD | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Structure / Year: 2025 Journal: Structure / Year: 2025Title: Structural diversity and oligomerization of bacterial ubiquitin-like proteins. Authors: Minheng Gong / Qiaozhen Ye / Yajie Gu / Lydia R Chambers / Andrey A Bobkov / Neal K Arakawa / Mariusz Matyszewski / Kevin D Corbett /  Abstract: Bacteria possess a variety of operons with homology to eukaryotic ubiquitination pathways that encode predicted E1, E2, E3, deubiquitinase, and ubiquitin-like proteins. Some of these pathways have ...Bacteria possess a variety of operons with homology to eukaryotic ubiquitination pathways that encode predicted E1, E2, E3, deubiquitinase, and ubiquitin-like proteins. Some of these pathways have recently been shown to function in anti-bacteriophage immunity, but the biological functions of others remain unknown. Here, we show that ubiquitin-like proteins in two bacterial operon families show surprising architectural diversity, possessing one to three β-grasp domains preceded by diverse N-terminal domains. We find that a large group of bacterial ubiquitin-like proteins possess three β-grasp domains and form homodimers and helical filaments mediated by conserved Ca ion binding sites. Our findings highlight a distinctive mode of self-assembly for ubiquitin-like proteins and suggest that Ca-mediated ubiquitin-like protein filament assembly and/or disassembly enables cells to sense and respond to stress conditions that alter intracellular metal ion concentration. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_46576.map.gz emd_46576.map.gz | 62.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-46576-v30.xml emd-46576-v30.xml emd-46576.xml emd-46576.xml | 19.4 KB 19.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_46576_fsc.xml emd_46576_fsc.xml | 10.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_46576.png emd_46576.png | 108.7 KB | ||
| Filedesc metadata |  emd-46576.cif.gz emd-46576.cif.gz | 6.2 KB | ||
| Others |  emd_46576_half_map_1.map.gz emd_46576_half_map_1.map.gz emd_46576_half_map_2.map.gz emd_46576_half_map_2.map.gz | 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-46576 http://ftp.pdbj.org/pub/emdb/structures/EMD-46576 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-46576 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-46576 | HTTPS FTP |
-Validation report
| Summary document |  emd_46576_validation.pdf.gz emd_46576_validation.pdf.gz | 873.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_46576_full_validation.pdf.gz emd_46576_full_validation.pdf.gz | 872.9 KB | Display | |
| Data in XML |  emd_46576_validation.xml.gz emd_46576_validation.xml.gz | 18.5 KB | Display | |
| Data in CIF |  emd_46576_validation.cif.gz emd_46576_validation.cif.gz | 23.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-46576 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-46576 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-46576 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-46576 | HTTPS FTP |
-Related structure data
| Related structure data |  9d59MC  9d5aC  9d5bC C: citing same article ( M: atomic model generated by this map |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_46576.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_46576.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.935 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_46576_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_46576_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Filament assembly of Citrobacter sp. ubiquitin-like (Ubl) protein...
| Entire | Name: Filament assembly of Citrobacter sp. ubiquitin-like (Ubl) protein with Ca ion |
|---|---|
| Components |
|
-Supramolecule #1: Filament assembly of Citrobacter sp. ubiquitin-like (Ubl) protein...
| Supramolecule | Name: Filament assembly of Citrobacter sp. ubiquitin-like (Ubl) protein with Ca ion type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Citrobacter sp. RHBSTW-00271 (bacteria) Citrobacter sp. RHBSTW-00271 (bacteria) |
| Molecular weight | Theoretical: 20 kDa/nm |
-Macromolecule #1: Multi-ubiquitin domain-containing protein
| Macromolecule | Name: Multi-ubiquitin domain-containing protein / type: protein_or_peptide / ID: 1 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Citrobacter sp. RHBSTW-00271 (bacteria) Citrobacter sp. RHBSTW-00271 (bacteria) |
| Molecular weight | Theoretical: 28.376852 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKSSHHHHHH ENLYFQSNAQ DIQSQHHHRR FIEVADETLS FRQVVMEDST PNGSQISAAS GFKPDQMPVV LMLLPNGSLE DIRPDEVVD LSSEVRRFIV VESDRTYFFT IDGARLEWPC RFITGYSIRQ LGDIGDNKKL LLEREDEADL EVQNDQIIDL D GDGIERFI ...String: MKSSHHHHHH ENLYFQSNAQ DIQSQHHHRR FIEVADETLS FRQVVMEDST PNGSQISAAS GFKPDQMPVV LMLLPNGSLE DIRPDEVVD LSSEVRRFIV VESDRTYFFT IDGARLEWPC RFITGYSIRQ LGDIGDNKKL LLEREDEADL EVQNDQIIDL D GDGIERFI SRKATWKLNI QGKEFTFDTP TVVIRDAVIR AGLNPNQAWH IFLKVEGQPK VEKNIDDVID LRTPGIEKLR LT PKDVNNG |
-Macromolecule #2: CALCIUM ION
| Macromolecule | Name: CALCIUM ION / type: ligand / ID: 2 / Number of copies: 14 / Formula: CA |
|---|---|
| Molecular weight | Theoretical: 40.078 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | helical reconstruction |
| Aggregation state | filament |
- Sample preparation
Sample preparation
| Buffer | pH: 8.5 Component:
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: PLASMA CLEANING / Pretreatment - Time: 10 sec. | ||||||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 298 K / Instrument: FEI VITROBOT MARK III |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 45.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.5 µm / Nominal magnification: 130000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)