[English] 日本語
 Yorodumi
Yorodumi- EMDB-4469: Spiral structure of E. coli RavA in the RavA-LdcI cage-like complex -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-4469 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Spiral structure of E. coli RavA in the RavA-LdcI cage-like complex | |||||||||||||||
 Map data Map data | Cryo-EM map of Class 1 of the E. coli RavA-LdcI cage-like complex after symmetry expansion and masked 3D classification. | |||||||||||||||
 Sample Sample |
| |||||||||||||||
 Keywords Keywords | Complex / MoxR ATPase / Lysine decarboxylase / HYDROLASE | |||||||||||||||
| Function / homology |  Function and homology information Function and homology informationarginine decarboxylase activity / lysine decarboxylase / lysine decarboxylase activity / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to catalyse transmembrane movement of substances / arginine catabolic process / pyridoxal phosphate binding / ATP hydrolysis activity / ATP binding / identical protein binding / cytosol / cytoplasm Similarity search - Function | |||||||||||||||
| Biological species |  | |||||||||||||||
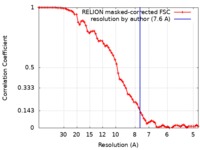
| Method | single particle reconstruction / cryo EM / Resolution: 7.6 Å | |||||||||||||||
 Authors Authors | Arragain B / Felix J | |||||||||||||||
| Funding support |  France, 4 items France, 4 items
| |||||||||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2020 Journal: Commun Biol / Year: 2020Title: Structural insights into ATP hydrolysis by the MoxR ATPase RavA and the LdcI-RavA cage-like complex. Authors: Matthew Jessop / Benoit Arragain / Roger Miras / Angélique Fraudeau / Karine Huard / Maria Bacia-Verloop / Patrice Catty / Jan Felix / Hélène Malet / Irina Gutsche /  Abstract: The hexameric MoxR AAA+ ATPase RavA and the decameric lysine decarboxylase LdcI form a 3.3 MDa cage, proposed to assist assembly of specific respiratory complexes in E. coli. Here, we show that ...The hexameric MoxR AAA+ ATPase RavA and the decameric lysine decarboxylase LdcI form a 3.3 MDa cage, proposed to assist assembly of specific respiratory complexes in E. coli. Here, we show that inside the LdcI-RavA cage, RavA hexamers adopt an asymmetric spiral conformation in which the nucleotide-free seam is constrained to two opposite orientations. Cryo-EM reconstructions of free RavA reveal two co-existing structural states: an asymmetric spiral, and a flat C2-symmetric closed ring characterised by two nucleotide-free seams. The closed ring RavA state bears close structural similarity to the pseudo two-fold symmetric crystal structure of the AAA+ unfoldase ClpX, suggesting a common ATPase mechanism. Based on these structures, and in light of the current knowledge regarding AAA+ ATPases, we propose different scenarios for the ATP hydrolysis cycle of free RavA and the LdcI-RavA cage-like complex, and extend the comparison to other AAA+ ATPases of clade 7. | |||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_4469.map.gz emd_4469.map.gz | 2.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-4469-v30.xml emd-4469-v30.xml emd-4469.xml emd-4469.xml | 27.3 KB 27.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_4469_fsc.xml emd_4469_fsc.xml | 7.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_4469.png emd_4469.png | 275.3 KB | ||
| Filedesc metadata |  emd-4469.cif.gz emd-4469.cif.gz | 8.6 KB | ||
| Others |  emd_4469_half_map_1.map.gz emd_4469_half_map_1.map.gz emd_4469_half_map_2.map.gz emd_4469_half_map_2.map.gz | 23.4 MB 23.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-4469 http://ftp.pdbj.org/pub/emdb/structures/EMD-4469 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4469 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-4469 | HTTPS FTP |
-Validation report
| Summary document |  emd_4469_validation.pdf.gz emd_4469_validation.pdf.gz | 799.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_4469_full_validation.pdf.gz emd_4469_full_validation.pdf.gz | 799 KB | Display | |
| Data in XML |  emd_4469_validation.xml.gz emd_4469_validation.xml.gz | 12.7 KB | Display | |
| Data in CIF |  emd_4469_validation.cif.gz emd_4469_validation.cif.gz | 17.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4469 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4469 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4469 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-4469 | HTTPS FTP |
-Related structure data
| Related structure data |  6q7lMC  4470C  6q7mC  6szaC  6szbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_4469.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_4469.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of Class 1 of the E. coli RavA-LdcI cage-like complex after symmetry expansion and masked 3D classification. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X: 2.42 Å / Y: 2.42 Å / Z: 2.42001 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
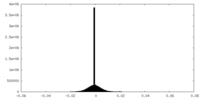
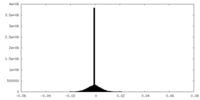
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
-Half map: Unfiltered half-map 1.
| File | emd_4469_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half-map 1. | ||||||||||||
| Projections & Slices |
| ||||||||||||
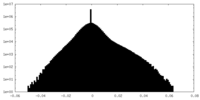
| Density Histograms |
-Half map: Unfiltered half-map 2.
| File | emd_4469_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unfiltered half-map 2. | ||||||||||||
| Projections & Slices |
| ||||||||||||
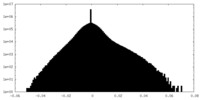
| Density Histograms |
- Sample components
Sample components
-Entire : Complex of the hexameric MoxR AAA+ ATPase RavA and the decameric ...
| Entire | Name: Complex of the hexameric MoxR AAA+ ATPase RavA and the decameric lysine decarboxylase LdcI. |
|---|---|
| Components |
|
-Supramolecule #1: Complex of the hexameric MoxR AAA+ ATPase RavA and the decameric ...
| Supramolecule | Name: Complex of the hexameric MoxR AAA+ ATPase RavA and the decameric lysine decarboxylase LdcI. type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 3.3 MDa |
-Macromolecule #1: Inducible lysine decarboxylase
| Macromolecule | Name: Inducible lysine decarboxylase / type: protein_or_peptide / ID: 1 / Number of copies: 20 / Enantiomer: LEVO / EC number: lysine decarboxylase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 81.357008 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNVIAILNHM GVYFKEEPIR ELHRALERLN FQIVYPNDRD DLLKLIENNA RLCGVIFDWD KYNLELCEEI SKMNENLPLY AFANTYSTL DVSLNDLRLQ ISFFEYALGA AEDIANKIKQ TTDEYINTIL PPLTKALFKY VREGKYTFCT PGHMGGTAFQ K SPVGSLFY ...String: MNVIAILNHM GVYFKEEPIR ELHRALERLN FQIVYPNDRD DLLKLIENNA RLCGVIFDWD KYNLELCEEI SKMNENLPLY AFANTYSTL DVSLNDLRLQ ISFFEYALGA AEDIANKIKQ TTDEYINTIL PPLTKALFKY VREGKYTFCT PGHMGGTAFQ K SPVGSLFY DFFGPNTMKS DISISVSELG SLLDHSGPHK EAEQYIARVF NADRSYMVTN GTSTANKIVG MYSAPAGSTI LI DRNCHKS LTHLMMMSDV TPIYFRPTRN AYGILGGIPQ SEFQHATIAK RVKETPNATW PVHAVITNST YDGLLYNTDF IKK TLDVKS IHFDSAWVPY TNFSPIYEGK CGMSGGRVEG KVIYETQSTH KLLAAFSQAS MIHVKGDVNE ETFNEAYMMH TTTS PHYGI VASTETAAAM MKGNAGKRLI NGSIERAIKF RKEIKRLRTE SDGWFFDVWQ PDHIDTTECW PLRSDSTWHG FKNID NEHM YLDPIKVTLL TPGMEKDGTM SDFGIPASIV AKYLDEHGIV VEKTGPYNLL FLFSIGIDKT KALSLLRALT DFKRAF DLN LRVKNMLPSL YREDPEFYEN MRIQELAQNI HKLIVHHNLP DLMYRAFEVL PTMVMTPYAA FQKELHGMTE EVYLDEM VG RINANMILPY PPGVPLVMPG EMITEESRPV LEFLQMLCEI GAHYPGFETD IHGAYRQADG RYTVKVLKEE SKK UniProtKB: Inducible lysine decarboxylase |
-Macromolecule #2: ATPase RavA
| Macromolecule | Name: ATPase RavA / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to catalyse transmembrane movement of substances |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 56.454586 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAHPHLLAER ISRLSSSLEK GLYERSHAIR LCLLAALSGE SVFLLGPPGI AKSLIARRLK FAFQNARAFE YLMTRFSTPE EVFGPLSIQ ALKDEGRYER LTSGYLPEAE IVFLDEIWKA GPAILNTLLT AINERQFRNG AHVEKIPMRL LVAASNELPE A DSSLEALY ...String: MAHPHLLAER ISRLSSSLEK GLYERSHAIR LCLLAALSGE SVFLLGPPGI AKSLIARRLK FAFQNARAFE YLMTRFSTPE EVFGPLSIQ ALKDEGRYER LTSGYLPEAE IVFLDEIWKA GPAILNTLLT AINERQFRNG AHVEKIPMRL LVAASNELPE A DSSLEALY DRMLIRLWLD KVQDKANFRS MLTSQQDEND NPVPDALQVT DEEYERWQKE IGEITLPDHV FELIFMLRQQ LD KLPDAPY VSDRRWKKAI RLLQASAFFS GRSAVAPVDL ILLKDCLWYD AQSLNLIQQQ IDVLMTGHAW QQQGMLTRLG AIV QRHLQL QQQQSDKTAL TVIRLGGIFS RRQQYQLPVN VTASTLTLLL QKPLKLHDME VVHISFERSA LEQWLSKGGE IRGK LNGIG FAQKLNLEVD SAQHLVVRDV SLQGSTLALP GSSAEGLPGE IKQQLEELES DWRKQHALFS EQQKCLFIPG DWLGR IEAS LQDVGAQIRQ AQQC UniProtKB: ATPase RavA |
-Macromolecule #3: ATPase RavA
| Macromolecule | Name: ATPase RavA / type: protein_or_peptide / ID: 3 / Number of copies: 5 / Enantiomer: LEVO EC number: Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to catalyse transmembrane movement of substances |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 56.351445 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAHPHLLAER ISRLSSSLEK GLYERSHAIR LCLLAALSGE SVFLLGPPGI AKSLIARRLK FAFQNARAFE YLMTRFSTPE EVFGPLSIQ ALKDEGRYER LTSGYLPEAE IVFLDEIWKA GPAILNTLLT AINERQFRNG AHVEKIPMRL LVAASNELPE A DSSLEALY ...String: MAHPHLLAER ISRLSSSLEK GLYERSHAIR LCLLAALSGE SVFLLGPPGI AKSLIARRLK FAFQNARAFE YLMTRFSTPE EVFGPLSIQ ALKDEGRYER LTSGYLPEAE IVFLDEIWKA GPAILNTLLT AINERQFRNG AHVEKIPMRL LVAASNELPE A DSSLEALY DRMLIRLWLD KVQDKANFRS MLTSQQDEND NPVPDALQVT DEEYERWQKE IGEITLPDHV FELIFMLRQQ LD KLPDAPY VSDRRWKKAI RLLQASAFFS GRSAVAPVDL ILLKDCLWYD AQSLNLIQQQ IDVLMTGHAW QQQGMLTRLG AIV QRHLQL QQQQSDKTAL TVIRLGGIFS RRQQYQLPVN VTASTLTLLL QKPLKLHDME VVHISFERSA LEQWLSKGGE IRGK LNGIG FAQKLNLEVD SAQHLVVRDV SLQGSTLALP GSSAEGLPGE IKQQLEELES DWRKQHALFS EQQKCLFIPG DWLGR IEAS LQDVGAQIRQ AQQ UniProtKB: ATPase RavA |
-Macromolecule #4: PYRIDOXAL-5'-PHOSPHATE
| Macromolecule | Name: PYRIDOXAL-5'-PHOSPHATE / type: ligand / ID: 4 / Number of copies: 20 / Formula: PLP |
|---|---|
| Molecular weight | Theoretical: 247.142 Da |
| Chemical component information |  ChemComp-PLP: |
-Macromolecule #5: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 5 / Number of copies: 5 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.98 mg/mL |
|---|---|
| Buffer | pH: 7.9 Details: 20 mM Tris pH 7.9, 300 mM NaCl, 2 mM ADP, 10 mM MgCl 2 , 0.1 mM PLP and 1 mM DTT |
| Grid | Model: Quantifoil, UltrAuFoil, R1.2/1.3 / Material: COPPER / Mesh: 400 / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE |
| Details | Concentrations of LdcI and RavA were 0.38 mg/ml (4.67 microM) and 0.6 mg/ml (10.64 microM) respectively. |
- Electron microscopy
Electron microscopy
| Microscope | FEI POLARA 300 |
|---|---|
| Details | Data collection was performed on an FEI Polara microscope operated at 300 kV. Movies of 40 frames were collected with a total exposure time of 8s and a total dose of 40e-/Angstrom^2 on a K2 summit direct electron detector (Gatan) at a magnification of 41270x, corresponding to 1.21 Angstrom/pixel at the specimen level. |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Digitization - Frames/image: 3-40 / Number real images: 1819 / Average exposure time: 8.0 sec. / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated magnification: 41270 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Tecnai Polara / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model |
| ||||||
|---|---|---|---|---|---|---|---|
| Details | Local resolution estimation and subsequent filtering of maps were performed in Relion3.0. For fitting of atomic models in the resulting filtered maps, we used the previously-determined X-ray structures of LdcI (PDB ID: 3N75) (Kanjee et al., 2011) and RavA (PDB ID: 3NBX) (El Bakkouri et al., 2010). In each map, two decameric LdcI molecules extracted from PDB 3N75 and one spiral RavA hexamer extracted from a continuous RavA helix generated from PDB 3NBX were fitted separately using iMODFIT (Lopez-Blanco and Chacon, 2013), followed by a single round of B-factor (ADP) refinement in Phenix. | ||||||
| Refinement | Protocol: FLEXIBLE FIT | ||||||
| Output model |  PDB-6q7l: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)