+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of S. cerevisiae PolE-core-DNA | ||||||||||||
 Map data Map data | structure of S. cerevisiae PolE-core-DNA | ||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Pol2 / DNA / REPLICATION / DNA BINDING PROTEIN-DNA complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationepsilon DNA polymerase complex / 4 iron, 4 sulfur cluster binding / DNA-directed DNA polymerase / DNA-directed DNA polymerase activity / DNA replication / DNA repair / nucleotide binding / DNA binding / zinc ion binding Similarity search - Function | ||||||||||||
| Biological species |  | ||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | ||||||||||||
 Authors Authors | Yuan Z / Georgescu R / O'Donnell M / Li H | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Science / Year: 2024 Journal: Science / Year: 2024Title: Mechanism of PCNA loading by Ctf18-RFC for leading-strand DNA synthesis. Authors: Zuanning Yuan / Roxana Georgescu / Nina Y Yao / Olga Yurieva / Michael E O'Donnell / Huilin Li /  Abstract: The proliferating cell nuclear antigen (PCNA) clamp encircles DNA to hold DNA polymerases (Pols) to DNA for processivity. The Ctf18-RFC PCNA loader, a replication factor C (RFC) variant, is specific ...The proliferating cell nuclear antigen (PCNA) clamp encircles DNA to hold DNA polymerases (Pols) to DNA for processivity. The Ctf18-RFC PCNA loader, a replication factor C (RFC) variant, is specific to the leading-strand Pol (Polε). We reveal here the underlying mechanism of Ctf18-RFC specificity to Polε using cryo-electron microscopy and biochemical studies. We found that both Ctf18-RFC and Polε contain specific structural features that direct PCNA loading onto DNA. Unlike other clamp loaders, Ctf18-RFC has a disordered ATPase associated with a diverse cellular activities (AAA+) motor that requires Polε to bind and stabilize it for efficient PCNA loading. In addition, Ctf18-RFC can pry prebound Polε off of DNA, then load PCNA onto DNA and transfer the PCNA-DNA back to Polε. These elements in both Ctf18-RFC and Polε provide specificity in loading PCNA onto DNA for Polε. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_44356.map.gz emd_44356.map.gz | 59.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-44356-v30.xml emd-44356-v30.xml emd-44356.xml emd-44356.xml | 16.8 KB 16.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_44356.png emd_44356.png | 185.3 KB | ||
| Filedesc metadata |  emd-44356.cif.gz emd-44356.cif.gz | 6.5 KB | ||
| Others |  emd_44356_half_map_1.map.gz emd_44356_half_map_1.map.gz emd_44356_half_map_2.map.gz emd_44356_half_map_2.map.gz | 59.3 MB 59.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-44356 http://ftp.pdbj.org/pub/emdb/structures/EMD-44356 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44356 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-44356 | HTTPS FTP |
-Validation report
| Summary document |  emd_44356_validation.pdf.gz emd_44356_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_44356_full_validation.pdf.gz emd_44356_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_44356_validation.xml.gz emd_44356_validation.xml.gz | 12.1 KB | Display | |
| Data in CIF |  emd_44356_validation.cif.gz emd_44356_validation.cif.gz | 14.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44356 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44356 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44356 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-44356 | HTTPS FTP |
-Related structure data
| Related structure data |  9b8rMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_44356.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_44356.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | structure of S. cerevisiae PolE-core-DNA | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.828 Å | ||||||||||||||||||||||||||||||||||||
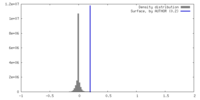
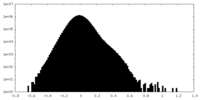
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half A map
| File | emd_44356_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half A map | ||||||||||||
| Projections & Slices |
| ||||||||||||
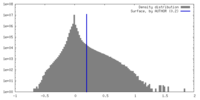
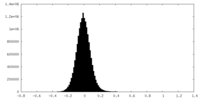
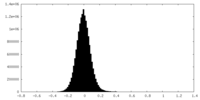
| Density Histograms |
-Half map: Half B map
| File | emd_44356_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half B map | ||||||||||||
| Projections & Slices |
| ||||||||||||
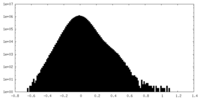
| Density Histograms |
- Sample components
Sample components
-Entire : PolE-core-DNA complex
| Entire | Name: PolE-core-DNA complex |
|---|---|
| Components |
|
-Supramolecule #1: PolE-core-DNA complex
| Supramolecule | Name: PolE-core-DNA complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Primer DNA
| Macromolecule | Name: Primer DNA / type: dna / ID: 1 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: DNA molecule (others) |
| Molecular weight | Theoretical: 3.026977 KDa |
| Sequence | String: (DC)(DT)(DG)(DT)(DT)(DG)(DC)(DT)(DG)(DC) |
-Macromolecule #2: Template DNA
| Macromolecule | Name: Template DNA / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: DNA molecule (others) |
| Molecular weight | Theoretical: 3.680432 KDa |
| Sequence | String: (DT)(DA)(DG)(DC)(DA)(DG)(DC)(DA)(DA)(DC) (DA)(DG) |
-Macromolecule #3: DNA polymerase epsilon catalytic subunit
| Macromolecule | Name: DNA polymerase epsilon catalytic subunit / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: DNA-directed DNA polymerase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 133.898031 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SNNYALSAQQ LLNASKIDDI DSMMGFERYV PPQYNGRFDA KDIDQIPGRV GWLTNMHATL VSQETLSSGS NGGGNSNDGE RVTTNQGIS GVDFYFLDEE GGSFKSTVVY DPYFFIACND ESRVNDVEEL VKKYLESCLK SLQIIRKEDL TMDNHLLGLQ K TLIKLSFV ...String: SNNYALSAQQ LLNASKIDDI DSMMGFERYV PPQYNGRFDA KDIDQIPGRV GWLTNMHATL VSQETLSSGS NGGGNSNDGE RVTTNQGIS GVDFYFLDEE GGSFKSTVVY DPYFFIACND ESRVNDVEEL VKKYLESCLK SLQIIRKEDL TMDNHLLGLQ K TLIKLSFV NSNQLFEARK LLRPILQDNA NNNVQRNIYN VAANGSEKVD AKHLIEDIRE YDVPYHVRVS IDKDIRVGKW YK VTQQGFI EDTRKIAFAD PVVMAFDIET TKPPLKFPDS AVDQIMMISY MIDGEGFLIT NREIISEDIE DFEYTPKPEY PGF FTIFNE NDEVALLQRF FEHIRDVRPT VISTFNGDFF DWPFIHNRSK IHGLDMFDEI GFAPDAEGEY KSSYCSHMDC FRWV KRDSY LPQGSQGLKA VTQSKLGYNP IELDPELMTP YAFEKPQHLS EYSVSDAVAT YYLYMKYVHP FIFSLCTIIP LNPDE TLRK GTGTLCEMLL MVQAYQHNIL LPNKHTDPIE RFYDGHLLES ETYVGGHVES LEAGVFRSDL KNEFKIDPSA IDELLQ ELP EALKFSVEVE NKSSVDKVTN FEEIKNQITQ KLLELKENNI RNELPLIYHV DVASMYPNIM TTNRLQPDSI KAERDCA SC DFNRPGKTCA RKLKWAWRGE FFPSKMDEYN MIKRALQNET FPNKNKFSKK KVLTFDELSY ADQVIHIKKR LTEYSRKV Y HRVKVSEIVE REAIVCQREN PFYVDTVKSF RDRRYEFKGL AKTWKGNLSK IDPSDKHARD EAKKMIVLYD SLQLAHKVI LNSFYGYVMR KGSRWYSMEM AGITCLTGAT IIQMARALVE RVGRPLELDT DGIWCILPKS FPETYFFTLE NGKKLYLSYP CSMLNYRVH QKFTNHQYQE LKDPLNYIYE THSENTIFFE VDGPYKAMIL PSSKEEGKGI KKRYAVFNED GSLAELKGFE L KRRGELQL IKNFQSDIFK VFLEGDTLEG CYSAVASVCN RWLDVLDSHG LMLEDEDLVS LICENRSMSK TLKEYEGQKS TS ITTARRL GDFLGEDMVK DKGLQCKYII SSKPFNAPVT ERAIPVAIFS ADIPIKRSFL RRWTLDPSLE DLDIRTIIDW GYY RERLGS AIQKIITIPA ALQGVSNPVP RVEHPDWLKR KIAT UniProtKB: DNA polymerase epsilon catalytic subunit |
-Macromolecule #4: IRON/SULFUR CLUSTER
| Macromolecule | Name: IRON/SULFUR CLUSTER / type: ligand / ID: 4 / Number of copies: 1 / Formula: SF4 |
|---|---|
| Molecular weight | Theoretical: 351.64 Da |
| Chemical component information |  ChemComp-FS1: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 121833 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)