[English] 日本語
 Yorodumi
Yorodumi- EMDB-43478: Structure of a synthetic antibody in complex with a class I MHC p... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of a synthetic antibody in complex with a class I MHC presenting a hapten-peptide conjugate | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Complex / Synthetic antibody / MHC class I / covalently modified peptide / IMMUNE SYSTEM | |||||||||
| Function / homology |  Function and homology information Function and homology informationforebrain astrocyte development / positive regulation of memory T cell activation / T cell mediated cytotoxicity directed against tumor cell target / TAP complex binding / negative regulation of epithelial cell differentiation / regulation of synaptic transmission, GABAergic / positive regulation of CD8-positive, alpha-beta T cell activation / CD8-positive, alpha-beta T cell activation / Golgi medial cisterna / positive regulation of CD8-positive, alpha-beta T cell proliferation ...forebrain astrocyte development / positive regulation of memory T cell activation / T cell mediated cytotoxicity directed against tumor cell target / TAP complex binding / negative regulation of epithelial cell differentiation / regulation of synaptic transmission, GABAergic / positive regulation of CD8-positive, alpha-beta T cell activation / CD8-positive, alpha-beta T cell activation / Golgi medial cisterna / positive regulation of CD8-positive, alpha-beta T cell proliferation / type I pneumocyte differentiation / epithelial tube branching involved in lung morphogenesis / CD8 receptor binding / Rac protein signal transduction / antigen processing and presentation of exogenous peptide antigen via MHC class I / skeletal muscle cell differentiation / positive regulation of Rac protein signal transduction / Signaling by RAS GAP mutants / Signaling by RAS GTPase mutants / Activation of RAS in B cells / endoplasmic reticulum exit site / RAS signaling downstream of NF1 loss-of-function variants / RUNX3 regulates p14-ARF / antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-dependent / TAP binding / SOS-mediated signalling / Activated NTRK3 signals through RAS / protection from natural killer cell mediated cytotoxicity / Activated NTRK2 signals through RAS / SHC1 events in ERBB4 signaling / Signalling to RAS / SHC-related events triggered by IGF1R / Activated NTRK2 signals through FRS2 and FRS3 / beta-2-microglobulin binding / glial cell proliferation / SHC-mediated cascade:FGFR2 / Estrogen-stimulated signaling through PRKCZ / SHC-mediated cascade:FGFR3 / MET activates RAS signaling / Signaling by PDGFRA transmembrane, juxtamembrane and kinase domain mutants / Signaling by PDGFRA extracellular domain mutants / PTK6 Regulates RHO GTPases, RAS GTPase and MAP kinases / T cell receptor binding / SHC-mediated cascade:FGFR4 / detection of bacterium / Signaling by FGFR4 in disease / Erythropoietin activates RAS / Signaling by CSF3 (G-CSF) / SHC-mediated cascade:FGFR1 / FRS-mediated FGFR2 signaling / protein-membrane adaptor activity / FRS-mediated FGFR3 signaling / Signaling by FLT3 ITD and TKD mutants / positive regulation of glial cell proliferation / Signaling by FGFR2 in disease / FRS-mediated FGFR4 signaling / p38MAPK events / Signaling by FGFR3 in disease / homeostasis of number of cells within a tissue / Tie2 Signaling / FRS-mediated FGFR1 signaling / striated muscle cell differentiation / GRB2 events in EGFR signaling / FLT3 Signaling / SHC1 events in EGFR signaling / EGFR Transactivation by Gastrin / Signaling by FLT3 fusion proteins / Signaling by FGFR1 in disease / GRB2 events in ERBB2 signaling / CD209 (DC-SIGN) signaling / Ras activation upon Ca2+ influx through NMDA receptor / NCAM signaling for neurite out-growth / SHC1 events in ERBB2 signaling / Downstream signal transduction / Constitutive Signaling by Overexpressed ERBB2 / Insulin receptor signalling cascade / Signaling by phosphorylated juxtamembrane, extracellular and kinase domain KIT mutants / positive regulation of ferrous iron binding / positive regulation of transferrin receptor binding / small monomeric GTPase / VEGFR2 mediated cell proliferation / positive regulation of receptor binding / early endosome lumen / Nef mediated downregulation of MHC class I complex cell surface expression / negative regulation of receptor binding / DAP12 interactions / antigen processing and presentation of endogenous peptide antigen via MHC class Ib / antigen processing and presentation of endogenous peptide antigen via MHC class I via ER pathway, TAP-independent / cellular response to iron ion / Endosomal/Vacuolar pathway / Antigen Presentation: Folding, assembly and peptide loading of class I MHC / lumenal side of endoplasmic reticulum membrane / FCERI mediated MAPK activation / Signaling by ERBB2 TMD/JMD mutants / RAF activation / cellular response to iron(III) ion / antigen processing and presentation of exogenous protein antigen via MHC class Ib, TAP-dependent / regulation of long-term neuronal synaptic plasticity / Constitutive Signaling by EGFRvIII / Signaling by high-kinase activity BRAF mutants Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
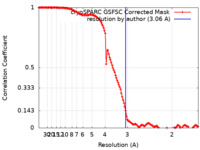
| Method | single particle reconstruction / cryo EM / Resolution: 3.06 Å | |||||||||
 Authors Authors | Maso L / Bang I / Koide S | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Molecular basis for antibody recognition of multiple drug-peptide/MHC complexes. Authors: Lorenzo Maso / Epsa Rajak / Injin Bang / Akiko Koide / Takamitsu Hattori / Benjamin G Neel / Shohei Koide /  Abstract: The HapImmune platform exploits covalent inhibitors as haptens for creating major histocompatibility complex (MHC)-presented tumor-specific neoantigens by design, combining targeted therapies with ...The HapImmune platform exploits covalent inhibitors as haptens for creating major histocompatibility complex (MHC)-presented tumor-specific neoantigens by design, combining targeted therapies with immunotherapy for the treatment of drug-resistant cancers. A HapImmune antibody, R023, recognizes multiple sotorasib-conjugated KRAS(G12C) peptides presented by different human leukocyte antigens (HLAs). This high specificity to sotorasib, coupled with broad HLA-binding capability, enables such antibodies, when reformatted as T cell engagers, to potently and selectively kill sotorasib-resistant KRAS(G12C) cancer cells expressing different HLAs upon sotorasib treatment. The loosening of HLA restriction could increase the patient population that can benefit from this therapeutic approach. To understand the molecular basis for its unconventional binding capability, we used single-particle cryogenic electron microscopy to determine the structures of R023 bound to multiple sotorasib-peptide conjugates presented by different HLAs. R023 forms a pocket for sotorasib between the V and V domains, binds HLAs in an unconventional, angled way, with V making most contacts with them, and makes few contacts with the peptide moieties. This binding mode enables the antibody to accommodate different hapten-peptide conjugates and to adjust its conformation to different HLAs presenting hapten-peptides. Deep mutational scanning validated the structures and revealed distinct levels of mutation tolerance by sotorasib- and HLA-binding residues. Together, our structural information and sequence landscape analysis reveal key features for achieving MHC-restricted recognition of multiple hapten-peptide antigens, which will inform the development of next-generation therapeutic antibodies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_43478.map.gz emd_43478.map.gz | 203.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-43478-v30.xml emd-43478-v30.xml emd-43478.xml emd-43478.xml | 22.4 KB 22.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_43478_fsc.xml emd_43478_fsc.xml | 12.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_43478.png emd_43478.png | 98.5 KB | ||
| Filedesc metadata |  emd-43478.cif.gz emd-43478.cif.gz | 6.9 KB | ||
| Others |  emd_43478_half_map_1.map.gz emd_43478_half_map_1.map.gz emd_43478_half_map_2.map.gz emd_43478_half_map_2.map.gz | 200.2 MB 200.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-43478 http://ftp.pdbj.org/pub/emdb/structures/EMD-43478 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43478 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-43478 | HTTPS FTP |
-Validation report
| Summary document |  emd_43478_validation.pdf.gz emd_43478_validation.pdf.gz | 978.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_43478_full_validation.pdf.gz emd_43478_full_validation.pdf.gz | 977.9 KB | Display | |
| Data in XML |  emd_43478_validation.xml.gz emd_43478_validation.xml.gz | 21.4 KB | Display | |
| Data in CIF |  emd_43478_validation.cif.gz emd_43478_validation.cif.gz | 27.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43478 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43478 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43478 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-43478 | HTTPS FTP |
-Related structure data
| Related structure data |  8vr9MC  8vraC  8vrbC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_43478.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_43478.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.825 Å | ||||||||||||||||||||||||||||||||||||
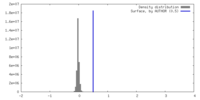
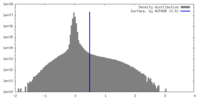
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: volume map half B
| File | emd_43478_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | volume_map_half_B | ||||||||||||
| Projections & Slices |
| ||||||||||||
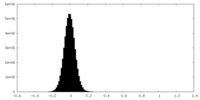
| Density Histograms |
-Half map: volume map half A
| File | emd_43478_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | volume_map_half_A | ||||||||||||
| Projections & Slices |
| ||||||||||||
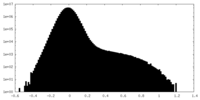
| Density Histograms |
- Sample components
Sample components
+Entire : Binary complex of Fab R023 with sotorasib-conjugated KRAS G12C pe...
+Supramolecule #1: Binary complex of Fab R023 with sotorasib-conjugated KRAS G12C pe...
+Supramolecule #2: Fab R023
+Supramolecule #3: class I MHC, having HLA-A*03:01, with Beta-2-microglobulin
+Supramolecule #4: sotorasib-conjugated KRAS G12C peptide (8-16)
+Macromolecule #1: HLA class I histocompatibility antigen, A alpha chain
+Macromolecule #2: Beta-2-microglobulin
+Macromolecule #3: GTPase KRas, N-terminally processed
+Macromolecule #4: R023 Fab light chain
+Macromolecule #5: R023 Fab heavy chain
+Macromolecule #6: AMG 510 (bound form)
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||||||
| Grid | Model: Quantifoil R0.6/1 / Material: GOLD / Mesh: 300 / Support film - Material: GOLD / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 80 sec. / Pretreatment - Atmosphere: AIR / Pretreatment - Pressure: 0.026000000000000002 kPa | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Average exposure time: 2.0 sec. / Average electron dose: 57.72 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 105000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)