[English] 日本語
 Yorodumi
Yorodumi- EMDB-42528: CryoEM structure of A/Perth/16/2009 H3 in complex with flu HA cen... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of A/Perth/16/2009 H3 in complex with flu HA central stem VH1-18 antibody UCA6 | |||||||||
 Map data Map data | Perth09H3 UCA6_sharp | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Flu Hemagglutinin / Central stem epitope / VH1-18 / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral budding from plasma membrane / clathrin-dependent endocytosis of virus by host cell / host cell surface receptor binding / apical plasma membrane / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Influenza A virus Influenza A virus | |||||||||
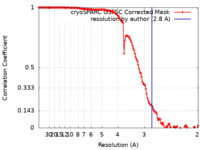
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Huang J / Han J / Ward AB | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Immunity / Year: 2024 Journal: Immunity / Year: 2024Title: Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses. Authors: Rashmi Ray / Faez Amokrane Nait Mohamed / Daniel P Maurer / Jiachen Huang / Berk A Alpay / Larance Ronsard / Zhenfei Xie / Julianna Han / Monica Fernandez-Quintero / Quynh Anh Phan / Rebecca ...Authors: Rashmi Ray / Faez Amokrane Nait Mohamed / Daniel P Maurer / Jiachen Huang / Berk A Alpay / Larance Ronsard / Zhenfei Xie / Julianna Han / Monica Fernandez-Quintero / Quynh Anh Phan / Rebecca L Ursin / Mya Vu / Kathrin H Kirsch / Thavaleak Prum / Victoria C Rosado / Thalia Bracamonte-Moreno / Vintus Okonkwo / Julia Bals / Caitlin McCarthy / Usha Nair / Masaru Kanekiyo / Andrew B Ward / Aaron G Schmidt / Facundo D Batista / Daniel Lingwood /   Abstract: Broadly neutralizing antibodies (bnAbs) targeting the hemagglutinin (HA) stem of influenza A viruses (IAVs) tend to be effective against either group 1 or group 2 viral diversity. In rarer cases, ...Broadly neutralizing antibodies (bnAbs) targeting the hemagglutinin (HA) stem of influenza A viruses (IAVs) tend to be effective against either group 1 or group 2 viral diversity. In rarer cases, intergroup protective bnAbs can be generated by human antibody paratopes that accommodate the conserved glycan differences between the group 1 and group 2 stems. We applied germline-engaging nanoparticle immunogens to elicit a class of cross-group bnAbs from physiological precursor frequency within a humanized mouse model. Cross-group protection depended on the presence of the human bnAb precursors within the B cell repertoire, and the vaccine-expanded antibodies enriched for an N55T substitution in the CDRH2 loop, a hallmark of the bnAb class. Structurally, this single mutation introduced a flexible fulcrum to accommodate glycosylation differences and could alone enable cross-group protection. Thus, broad IAV immunity can be expanded from the germline repertoire via minimal antigenic input and an exceptionally simple antibody development pathway. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42528.map.gz emd_42528.map.gz | 204 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42528-v30.xml emd-42528-v30.xml emd-42528.xml emd-42528.xml | 19.1 KB 19.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42528_fsc.xml emd_42528_fsc.xml | 12.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_42528.png emd_42528.png | 92 KB | ||
| Filedesc metadata |  emd-42528.cif.gz emd-42528.cif.gz | 6.4 KB | ||
| Others |  emd_42528_half_map_1.map.gz emd_42528_half_map_1.map.gz emd_42528_half_map_2.map.gz emd_42528_half_map_2.map.gz | 200.1 MB 200.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42528 http://ftp.pdbj.org/pub/emdb/structures/EMD-42528 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42528 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42528 | HTTPS FTP |
-Validation report
| Summary document |  emd_42528_validation.pdf.gz emd_42528_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42528_full_validation.pdf.gz emd_42528_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_42528_validation.xml.gz emd_42528_validation.xml.gz | 21.5 KB | Display | |
| Data in CIF |  emd_42528_validation.cif.gz emd_42528_validation.cif.gz | 27.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42528 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42528 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42528 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42528 | HTTPS FTP |
-Related structure data
| Related structure data |  8ut3MC  8ut4C  8ut5C  8ut6C  8ut7C  8ut8C  8ut9C  8uwaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42528.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42528.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Perth09H3 UCA6_sharp | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.982 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Perth09H3 UCA6 halfB
| File | emd_42528_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Perth09H3 UCA6_halfB | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Perth09H3 UCA6 halfA
| File | emd_42528_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Perth09H3 UCA6_halfA | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : An UCA6 Fab complex with one protomer of a H3 trimer
| Entire | Name: An UCA6 Fab complex with one protomer of a H3 trimer |
|---|---|
| Components |
|
-Supramolecule #1: An UCA6 Fab complex with one protomer of a H3 trimer
| Supramolecule | Name: An UCA6 Fab complex with one protomer of a H3 trimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: UCA6 HC Fv
| Macromolecule | Name: UCA6 HC Fv / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 13.906533 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QVQLVQSGAE VKKPGASVKV SCKASGYTFT SYGISWVRQA PGQGLEWMGW ISAYNGNTNY AQKLQGRVTM TTDTSTSTAY MELRSLRSD DTAVYYCARG LLQGVVILDS YYYTMDVWGQ GTTVTVSS |
-Macromolecule #2: UCA6 LC Fv
| Macromolecule | Name: UCA6 LC Fv / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 11.858149 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EIVLTQSPGT LSLSPGERAT LSCRASQSVS SSYLAWYQQK PGQAPRLLIY GASSRATGIP DRFSGSGSGT DFTLTISRLE PEDFAVYYC QQYGSSPRWT FGQGTKLEIK |
-Macromolecule #3: Hemagglutinin
| Macromolecule | Name: Hemagglutinin / type: protein_or_peptide / ID: 3 / Details: A/Perth/16/2009 H3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Influenza A virus Influenza A virus |
| Molecular weight | Theoretical: 62.717074 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QKLPGNDNST ATLCLGHHAV PNGTIVKTIT NDQIEVTNAT ELVQSSSTGE ICDSPHQILD GKNCTLIDAL LGDPQCDGFQ NKKWDLFVE RSKAYSNCYP YDVPDYASLR SLVASSGTLE FNNESFNWTG VTQNGTSSAC IRRSKNSFFS RLNWLTHLNF K YPALNVTM ...String: QKLPGNDNST ATLCLGHHAV PNGTIVKTIT NDQIEVTNAT ELVQSSSTGE ICDSPHQILD GKNCTLIDAL LGDPQCDGFQ NKKWDLFVE RSKAYSNCYP YDVPDYASLR SLVASSGTLE FNNESFNWTG VTQNGTSSAC IRRSKNSFFS RLNWLTHLNF K YPALNVTM PNNEQFDKLY IWGVHHPGTD KDQIFLYAQA SGRITVSTKR SQQTVSPNIG SRPRVRNIPS RISIYWTIVK PG DILLINS TGNLIAPRGY FKIRSGKSSI MRSDAPIGKC NSECITPNGS IPNDKPFQNV NRITYGACPR YVKQNTLKLA TGM RNVPEK QTRGIFGAIA GFIENGWEGM VDGWYGFRHQ NSEGRGQAAD LKSTQAAIDQ INGKLNRLIG KTNEKFHQIE KEFS EVEGR IQDLEKYVED TKIDLWSYNA ELLVALENQH TIDLTDSEMN KLFEKTKKQL RENAEDMGNG CFKIYHKCDN ACIGS IRNG TYDHDVYRDE ALNNRFQIKG GSGYIPEAPR DGQAYVRKDG EWVLLSTFLG SGLNDIFEAQ KIEWHEGHHH HHH UniProtKB: Hemagglutinin |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 21 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.7 mg/mL | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| ||||||||
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. | ||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.7000000000000001 µm |
 Movie
Movie Controller
Controller
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)