+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of the Clostridium thermocellum AdhE spirosome | |||||||||
 Map data Map data | Map of the AdhE spirosome | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Aldehyde-alcohol dehydrogenase / Complex / Spirosome / OXIDOREDUCTASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationbutanol dehydrogenase (NAD+) activity / acetaldehyde dehydrogenase (acetylating) activity / alcohol metabolic process / carbon utilization / metal ion binding Similarity search - Function | |||||||||
| Biological species |  Acetivibrio thermocellus DSM 1313 (bacteria) Acetivibrio thermocellus DSM 1313 (bacteria) | |||||||||
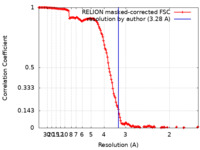
| Method | single particle reconstruction / cryo EM / Resolution: 3.28 Å | |||||||||
 Authors Authors | Ziegler SJ / Gruber JN | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2024 Journal: Elife / Year: 2024Title: Structural characterization and dynamics of AdhE ultrastructures from Clostridium thermocellum: A containment strategy for toxic intermediates Authors: Ziegler S / Knott B / Gruber J / Hengge N / Xu Q / Olson D / Romero E / Joubert L / Bomble Y | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42284.map.gz emd_42284.map.gz | 249.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42284-v30.xml emd-42284-v30.xml emd-42284.xml emd-42284.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_42284_fsc.xml emd_42284_fsc.xml | 14.7 KB | Display |  FSC data file FSC data file |
| Images |  emd_42284.png emd_42284.png | 74.1 KB | ||
| Masks |  emd_42284_msk_1.map emd_42284_msk_1.map | 266.8 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-42284.cif.gz emd-42284.cif.gz | 6.3 KB | ||
| Others |  emd_42284_half_map_1.map.gz emd_42284_half_map_1.map.gz emd_42284_half_map_2.map.gz emd_42284_half_map_2.map.gz | 212.7 MB 213.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42284 http://ftp.pdbj.org/pub/emdb/structures/EMD-42284 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42284 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42284 | HTTPS FTP |
-Validation report
| Summary document |  emd_42284_validation.pdf.gz emd_42284_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42284_full_validation.pdf.gz emd_42284_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_42284_validation.xml.gz emd_42284_validation.xml.gz | 21.9 KB | Display | |
| Data in CIF |  emd_42284_validation.cif.gz emd_42284_validation.cif.gz | 28.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42284 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42284 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42284 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42284 | HTTPS FTP |
-Related structure data
| Related structure data |  8uhwMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_42284.map.gz / Format: CCP4 / Size: 266.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42284.map.gz / Format: CCP4 / Size: 266.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map of the AdhE spirosome | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
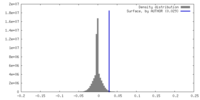
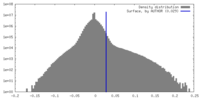
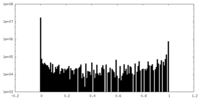
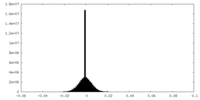
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_42284_msk_1.map emd_42284_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 1
| File | emd_42284_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half map 2
| File | emd_42284_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half map 2 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Homo-oligomer complex of AdhE
| Entire | Name: Homo-oligomer complex of AdhE |
|---|---|
| Components |
|
-Supramolecule #1: Homo-oligomer complex of AdhE
| Supramolecule | Name: Homo-oligomer complex of AdhE / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Acetivibrio thermocellus DSM 1313 (bacteria) Acetivibrio thermocellus DSM 1313 (bacteria) |
-Macromolecule #1: Aldehyde-alcohol dehydrogenase
| Macromolecule | Name: Aldehyde-alcohol dehydrogenase / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Acetivibrio thermocellus DSM 1313 (bacteria) Acetivibrio thermocellus DSM 1313 (bacteria) |
| Molecular weight | Theoretical: 97.93868 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHENL YFQGMTKIAN KYEVIDNVEK LEKALKRLRE AQSVYATYTQ EQVDKIFFEA AMAANKMRIP LAKMAVEETG MGVVEDKVI KNHYASEYIY NAYKNTKTCG VIEEDPAFGI KKIAEPLGVI AAVIPTTNPT STAIFKTLIA LKTRNAIIIS P HPRAKNST ...String: MHHHHHHENL YFQGMTKIAN KYEVIDNVEK LEKALKRLRE AQSVYATYTQ EQVDKIFFEA AMAANKMRIP LAKMAVEETG MGVVEDKVI KNHYASEYIY NAYKNTKTCG VIEEDPAFGI KKIAEPLGVI AAVIPTTNPT STAIFKTLIA LKTRNAIIIS P HPRAKNST IEAAKIVLEA AVKAGAPEGI IGWIDVPSLE LTNLVMREAD VILATGGPGL VKAAYSSGKP AIGVGAGNTP AI IDDSADI VLAVNSIIHS KTFDNGMICA SEQSVIVLDG VYKEVKKEFE KRGCYFLNED ETEKVRKTII INGALNAKIV GQK AHTIAN LAGFEVPETT KILIGEVTSV DISEEFAHEK LCPVLAMYRA KDFDDALDKA ERLVADGGFG HTSSLYIDTV TQKE KLQKF SERMKTCRIL VNTPSSQGGI GDLYNFKLAP SLTLGCGSWG GNSVSDNVGV KHLLNIKTVA ERRENMLWFR TPEKI YIKR GCLPVALDEL KNVMGKKKAF IVTDNFLYNN GYTKPITDKL DEMGIVHKTF FDVSPDPSLA SAKAGAAEML AFQPDT IIA VGGGSAMDAA KIMWVMYEHP EVDFMDMAMR FMDIRKRVYT FPKMGQKAYF IAIPTSAGTG SEVTPFAVIT DEKTGIK YP LADYELLPDM AIVDADMMMN APKGLTAASG IDALTHALEA YVSMLATDYT DSLALRAIKM IFEYLPRAYE NGASDPVA R EKMANAATIA GMAFANAFLG VCHSMAHKLG AFYHLPHGVA NALMINEVIR FNSSEAPTKM GTFPQYDHPR TLERYAEIA DYIGLKGKNN EEKVENLIKA IDELKEKVGI RKTIKDYDID EKEFLDRLDE MVEQAFDDQC TGTNPRYPLM NEIRQMYLNA YYGGAKK UniProtKB: Aldehyde-alcohol dehydrogenase |
-Macromolecule #2: FE (III) ION
| Macromolecule | Name: FE (III) ION / type: ligand / ID: 2 / Number of copies: 5 / Formula: FE |
|---|---|
| Molecular weight | Theoretical: 55.845 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.4 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: HOLEY / Support film - Film thickness: 20 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 10 sec. | |||||||||
| Vitrification | Cryogen name: ETHANE / Instrument: GATAN CRYOPLUNGE 3 |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|---|
| Output model |  PDB-8uhw: |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)