+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of Sec7 autoinhibited conformation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GEF / Exchange Factor / Golgi / PROTEIN TRANSPORT | |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of ARF protein signal transduction / guanyl-nucleotide exchange factor activity / protein transport / Golgi apparatus Similarity search - Function | |||||||||
| Biological species |  Thermothielavioides terrestris (fungus) Thermothielavioides terrestris (fungus) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.7 Å | |||||||||
 Authors Authors | Brownfield BA / Fromme JC | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2024 Journal: Proc Natl Acad Sci U S A / Year: 2024Title: Sec7 regulatory domains scaffold autoinhibited and active conformations. Authors: Bryce A Brownfield / Brian C Richardson / Steve L Halaby / J Christopher Fromme /  Abstract: The late stages of Golgi maturation involve a series of sequential trafficking events in which cargo-laden vesicles are produced and targeted to multiple distinct subcellular destinations. Each of ...The late stages of Golgi maturation involve a series of sequential trafficking events in which cargo-laden vesicles are produced and targeted to multiple distinct subcellular destinations. Each of these vesicle biogenesis events requires activation of an Arf GTPase by the Sec7/BIG guanine nucleotide exchange factor (GEF). Sec7 localization and activity is regulated by autoinhibition, positive feedback, and interaction with other GTPases. Although these mechanisms have been characterized biochemically, we lack a clear picture of how GEF localization and activity is modulated by these signals. Here, we report the cryogenic electron microscopy structure of full-length Sec7 in its autoinhibited form, revealing the architecture of its multiple regulatory domains. We use functional experiments to determine the basis for autoinhibition and use structural predictions to produce a model for an active conformation of the GEF that is supported empirically. This study therefore elucidates the conformational transition that Sec7 undergoes to become active on the organelle membrane surface. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_42135.map.gz emd_42135.map.gz | 8.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-42135-v30.xml emd-42135-v30.xml emd-42135.xml emd-42135.xml | 19.5 KB 19.5 KB | Display Display |  EMDB header EMDB header |
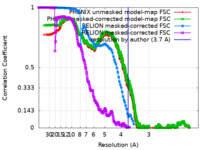
| FSC (resolution estimation) |  emd_42135_fsc.xml emd_42135_fsc.xml emd_42135_fsc_2.xml emd_42135_fsc_2.xml emd_42135_fsc_3.xml emd_42135_fsc_3.xml emd_42135_fsc_4.xml emd_42135_fsc_4.xml | 715.7 KB 715.3 KB 7.7 KB 16.9 KB | Display Display Display Display |  FSC data file FSC data file |
| Images |  emd_42135.png emd_42135.png | 26.7 KB | ||
| Filedesc metadata |  emd-42135.cif.gz emd-42135.cif.gz | 7.4 KB | ||
| Others |  emd_42135_additional_1.map.gz emd_42135_additional_1.map.gz emd_42135_additional_2.map.gz emd_42135_additional_2.map.gz | 1.2 MB 1.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-42135 http://ftp.pdbj.org/pub/emdb/structures/EMD-42135 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42135 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-42135 | HTTPS FTP |
-Validation report
| Summary document |  emd_42135_validation.pdf.gz emd_42135_validation.pdf.gz | 327.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_42135_full_validation.pdf.gz emd_42135_full_validation.pdf.gz | 327.1 KB | Display | |
| Data in XML |  emd_42135_validation.xml.gz emd_42135_validation.xml.gz | 90.7 KB | Display | |
| Data in CIF |  emd_42135_validation.cif.gz emd_42135_validation.cif.gz | 234.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42135 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42135 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42135 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-42135 | HTTPS FTP |
-Related structure data
| Related structure data |  8ucqMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_42135.map.gz / Format: CCP4 / Size: 440.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_42135.map.gz / Format: CCP4 / Size: 440.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.7415 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Additional map: #2
| File | emd_42135_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Additional map: #1
| File | emd_42135_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Sec7 homodimer
| Entire | Name: Sec7 homodimer |
|---|---|
| Components |
|
-Supramolecule #1: Sec7 homodimer
| Supramolecule | Name: Sec7 homodimer / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Thermothielavioides terrestris (fungus) Thermothielavioides terrestris (fungus) |
| Molecular weight | Theoretical: 392.14 KDa |
-Macromolecule #1: SEC7 domain-containing protein
| Macromolecule | Name: SEC7 domain-containing protein / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Thermothielavioides terrestris (fungus) Thermothielavioides terrestris (fungus) |
| Molecular weight | Theoretical: 215.563719 KDa |
| Recombinant expression | Organism:  Komagataella pastoris (fungus) Komagataella pastoris (fungus) |
| Sequence | String: MEQKLISEED LNSAVDHHHH HHRIPGLINS SLKFVVSSLD IIAAQAGRNK QLAELAEKAL AAIKENDQQL PDPEVVFAPL QLATKSGTI PLTTTALDCI GKLISYSYFS APSSSATQDG TEQTPLIERA IDTICDCFQG ETTLVEIQLQ IVKSLLAAVL N DKIIVHGA ...String: MEQKLISEED LNSAVDHHHH HHRIPGLINS SLKFVVSSLD IIAAQAGRNK QLAELAEKAL AAIKENDQQL PDPEVVFAPL QLATKSGTI PLTTTALDCI GKLISYSYFS APSSSATQDG TEQTPLIERA IDTICDCFQG ETTLVEIQLQ IVKSLLAAVL N DKIIVHGA GLLKAVRQVY NIFLLSRSTA NQQVAQGTLT QMVGTVFERV KTRLHMKEAR ANLGRLKASR SSLAVDRSDD QD SQAGKVD GEDATVETVS DATPSESVDK AGGGKLTLKD LEHRKSFDDS HMGDGPTMVS QVKPMKKASR SVSEQSLQES PQD ETPESL DAEDEAYIRD AYLVFRSFCN LSTKILPPDQ LYDLRGQPMR SKLISLHLIH TLLNNHITVF TSPLCTIRNT KNNE PTNFL QAIKYYLCLS ITRNGASSVD RVFDICCEIF WLMLKYMRSS FKNEIEVFLN EIYLALLARR NAPLSQKLTF VGILK RLCE DPRALVELYL NYDCNQNVDN IFQTIVEDLS RFATASVPIT PTQEQQYEES RSKSATAGEW QIKSVLPPPL SVALIA TNH EADTELPKEY VMKRTALDSL VETLRSLVHW SQPGRPELNG ASGDVQRRTS SDDLGDSIDP SMSETASRME VPIAPAT PV IDDDPDQLEK EKARKTAMTN AIKVFNFKPK HGIKLLIKEG FIPSDKPEDI ARFLLREERL DKAQIGEYLG EGDQKNVD I MHAFVDMMDF SKKRFVDALR EFLQAFRLPG EAQKIDRFML KFAHRYVTGN PNAFANADTP YVLAYSVIML NTDLHSSKV VKRMSKAEFI KNNRGINDNA DLPDEYLIGI YDDIASNEIV LKSEREAAAA AGTLPAQSTG LAGLGQAFSN VGRDLQREAY VQQSEEISL RSEQLFRDLY RSQRKSATKG GVKFISATSF KHVGPMFDAT WMSFFSTLSS LVQKTHNLDV NKLCLEGMKL A TKIACLFD LSTPREAFIS MLKNTANLNN PREMQAKNVE ALKVLLDLAQ TEGNYLKESW KDVLLCISQL DRLQLISGGV DE SAVPDVS RARFVPPPRT ETGESRKSTS SARRTRPRAH TGPQGVSLEI ALESRSDEVI KSVDRIFTNT ANLSRDAIIH FAR ALTEVS WDEIKVSGSN DSPRTYSLQK IVEISYYNMT RVRFEWSHIW DVLGEHFNRV GCHANTAIVF FALDSLRQLS MRFM EIEEL AGFKFQKDFL KPFEHVMSNS SNVTVKDMVL RCLIQMIQAR GENIRSGWRT MFGVFTVAAR EPYESIVNLA YENVT QVYK TRFGVVISQG AFTDLIVCLT EFSKNMRFQK KSLQAMETLK SVIPTMLKTP ECPLSQHKPT ATTASGSESH SKKAAV QQT RTSVEEGFWF PVLFAFHDVL MTGEDLEVRS NALNYFFETL LRYGGDFPPE FWDILWRQQL YPIFMVLRSR PEMTNAL NH EELSVWLSTT MIQALRNMIT LFTHYFDALE YMLDRFLELL ALCICQENDT IARIGSNCLQ QLILQNVTKF TAEHWAKI V GAFCELFERT TAYQLFSATT INSTASLSPP PSGLELGGPL SPTSATAPVD GKSLKINGVE TNGQTPGAEP ANGDADGNG TAAAAADASA PAATPQPQQG PAQQLEEFKP NNPLQQQPVV VTAARRRFFN RIISRCVLQL LMIETVNELF SNDAVYAQIP SAELLRLMA LLKKSFLFAK RFNADKDLRM RLWREGFMKQ PPNLLKQESG SAATYVAILF RMFGDTAPDR RGSRADVEAA L VPLCRDII RGYTALDDES QHRNIVAWRP VVVDVLEGYA AFPRDAFAAH IRSFYPLVVE LLGKDLGQDL RAALLLVLRR VG EVGLGIE GMGSGGAAAA AAAGAAAASS GQGNGNGAAA AAADSERRSS VLSVPSGPRH TPSMDSLNDD PSRQVMGKAE QKL ISEEDL NSAVDHHHHH H UniProtKB: A7bd1859-5d22-4ce8-aff3-d8d273e6acf0 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 8.0 mg/mL | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: OTHER | |||||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277.15 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Specialist optics | Energy filter - Name: GIF Bioquantum / Energy filter - Slit width: 20 eV |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number grids imaged: 1 / Number real images: 4474 / Average exposure time: 3.092 sec. / Average electron dose: 55.6 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 50.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 63000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Details | Phenix Real Space Refinement |
| Refinement | Space: REAL / Protocol: OTHER |
| Output model |  PDB-8ucq: |
 Movie
Movie Controller
Controller






 X (Sec.)
X (Sec.) Y (Row.)
Y (Row.) Z (Col.)
Z (Col.)