[English] 日本語
 Yorodumi
Yorodumi- EMDB-41874: CryoEM structure of A/Solomon Islands/3/2006 H1 HA in complex wit... -
+ Open data
Open data
- Basic information
Basic information
| Entry | 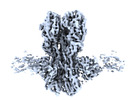 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of A/Solomon Islands/3/2006 H1 HA in complex with 05.GC.w2.3C10-H1_SI06 | |||||||||
 Map data Map data | CryoEM map of A/Solomon Islands/3/2006 H1 HA in complex with 3C10 mAb | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Influenza virus / hemagglutinin / H1 / monoclonal antibody / virus / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationviral budding from plasma membrane / clathrin-dependent endocytosis of virus by host cell / host cell surface receptor binding / apical plasma membrane / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   Influenza A virus Influenza A virus | |||||||||
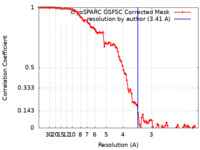
| Method | single particle reconstruction / cryo EM / Resolution: 3.41 Å | |||||||||
 Authors Authors | Moore N / Han J / Ward AB / Wilson IA | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Persistence of germinal center B cell responses after influenza virus vaccination in humans Authors: McIntire KM / Meng H / Lin TH / Kim W / Moore NE / Han J / McMahon M / Wang M / Malladi SK / Mohammed BM / Zhou JQ / Schmitz AJ / Hoehn K / Suessen T / Middleton WD / Teefey SA / Presti RM / ...Authors: McIntire KM / Meng H / Lin TH / Kim W / Moore NE / Han J / McMahon M / Wang M / Malladi SK / Mohammed BM / Zhou JQ / Schmitz AJ / Hoehn K / Suessen T / Middleton WD / Teefey SA / Presti RM / Krammer F / Turner JS / Ward AB / Wilson IA / Kleinstein SH / Ellebedy AH | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41874.map.gz emd_41874.map.gz | 86.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41874-v30.xml emd-41874-v30.xml emd-41874.xml emd-41874.xml | 21.2 KB 21.2 KB | Display Display |  EMDB header EMDB header |
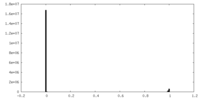
| FSC (resolution estimation) |  emd_41874_fsc.xml emd_41874_fsc.xml | 9.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_41874.png emd_41874.png | 88.1 KB | ||
| Masks |  emd_41874_msk_1.map emd_41874_msk_1.map | 91.1 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-41874.cif.gz emd-41874.cif.gz | 6.8 KB | ||
| Others |  emd_41874_half_map_1.map.gz emd_41874_half_map_1.map.gz emd_41874_half_map_2.map.gz emd_41874_half_map_2.map.gz | 84.6 MB 84.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41874 http://ftp.pdbj.org/pub/emdb/structures/EMD-41874 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41874 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41874 | HTTPS FTP |
-Validation report
| Summary document |  emd_41874_validation.pdf.gz emd_41874_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41874_full_validation.pdf.gz emd_41874_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_41874_validation.xml.gz emd_41874_validation.xml.gz | 17.4 KB | Display | |
| Data in CIF |  emd_41874_validation.cif.gz emd_41874_validation.cif.gz | 22.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41874 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41874 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41874 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41874 | HTTPS FTP |
-Related structure data
| Related structure data |  8u44MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41874.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41874.map.gz / Format: CCP4 / Size: 91.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
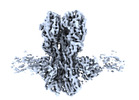
| Annotation | CryoEM map of A/Solomon Islands/3/2006 H1 HA in complex with 3C10 mAb | ||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||
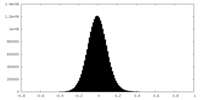
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_41874_msk_1.map emd_41874_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
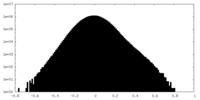
| Projections & Slices |
| ||||||||||||
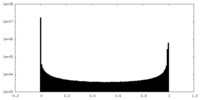
| Density Histograms |
-Half map: CryoEM half map A of A/Solomon Islands/3/2006 H1...
| File | emd_41874_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM half map A of A/Solomon Islands/3/2006 H1 HA in complex with 3C10 mAb | ||||||||||||
| Projections & Slices |
| ||||||||||||
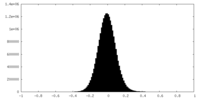
| Density Histograms |
-Half map: CryoEM half map B of A/Solomon Islands/3/2006 H1...
| File | emd_41874_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | CryoEM half map B of A/Solomon Islands/3/2006 H1 HA in complex with 3C10 mAb | ||||||||||||
| Projections & Slices |
| ||||||||||||
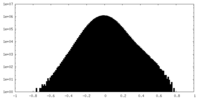
| Density Histograms |
- Sample components
Sample components
-Entire : Immune complex of A/Solomon Islands/3/2006 H1 HA with Fab 3C10
| Entire | Name: Immune complex of A/Solomon Islands/3/2006 H1 HA with Fab 3C10 |
|---|---|
| Components |
|
-Supramolecule #1: Immune complex of A/Solomon Islands/3/2006 H1 HA with Fab 3C10
| Supramolecule | Name: Immune complex of A/Solomon Islands/3/2006 H1 HA with Fab 3C10 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #3-#4, #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #2: Fab
| Supramolecule | Name: Fab / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: HA
| Supramolecule | Name: HA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3-#4 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: 05.GC.w2.3C10-H1_SI06 Heavy chain
| Macromolecule | Name: 05.GC.w2.3C10-H1_SI06 Heavy chain / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.771057 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: METDTLLLWV LLLWVPGSTG DEVQLVQSGA EVKKPGSSVR VSCKASGGTF SAISWVRQAP GQGLEWMGGI IPVFGTANYA QKFQGRVTI TADDSTSTAY MEVSSLRSDD TAVYYCAREE TWKGATIGVM GIWGQGTMVT VSSASTKGPS VFPLAPSSKS T SGGTAALG ...String: METDTLLLWV LLLWVPGSTG DEVQLVQSGA EVKKPGSSVR VSCKASGGTF SAISWVRQAP GQGLEWMGGI IPVFGTANYA QKFQGRVTI TADDSTSTAY MEVSSLRSDD TAVYYCAREE TWKGATIGVM GIWGQGTMVT VSSASTKGPS VFPLAPSSKS T SGGTAALG CLVKDYFPEP VTVSWNSGAL TSGVHTFPAV LQSSGLYSLS SVVTVPSSSL GTQTYICNVN HKPSNTKVDK RV EPKSC |
-Macromolecule #2: 05.GC.w2.3C10-H1_SI06 Light chain
| Macromolecule | Name: 05.GC.w2.3C10-H1_SI06 Light chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 25.706625 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: METDTLLLWV LLLWVPGSTG DDIQMTQSPS SLSASVGDRV TITCRASQSI SSYLNWYQHK PGKAPKLLIF TASNLQSGVP SRFSGGGSG TDFTLTISSL QPEDFATYYC QQSYTSPRTF GQGTKVEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN F YPREAKVQ ...String: METDTLLLWV LLLWVPGSTG DDIQMTQSPS SLSASVGDRV TITCRASQSI SSYLNWYQHK PGKAPKLLIF TASNLQSGVP SRFSGGGSG TDFTLTISSL QPEDFATYYC QQSYTSPRTF GQGTKVEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN F YPREAKVQ WKVDNALQSG NSQESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #3: Hemagglutinin HA1 chain
| Macromolecule | Name: Hemagglutinin HA1 chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Influenza A virus Influenza A virus |
| Molecular weight | Theoretical: 40.835902 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MVLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFAAD PGDTICIGYH ANNSTDTVDT VLEKNVTVTH SVNLLEDSHN GKLCRLKGI APLQLGNCSV AGWILGNPEC ELLISRESWS YIVEKPNPEN GTCYPGHFAD YEELREQLSS VSSFERFEIF P KESSWPNH ...String: MVLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFAAD PGDTICIGYH ANNSTDTVDT VLEKNVTVTH SVNLLEDSHN GKLCRLKGI APLQLGNCSV AGWILGNPEC ELLISRESWS YIVEKPNPEN GTCYPGHFAD YEELREQLSS VSSFERFEIF P KESSWPNH TTTGVSASCS HNGESSFYKN LLWLTGKNGL YPNLSKSYAN NKEKEVLVLW GVHHPPNIGD QRALYHKENA YV SVVSSHY SRKFTPEIAK RPKVRDQEGR INYYWTLLEP GDTIIFEANG NLIAPRYAFA LSRGFGSGII NSNAPMDECD AKC QTPQGA INSSLPFQNV HPVTIGECPK YVRSAKLRMV TGLRNIPSIQ SR UniProtKB: Hemagglutinin |
-Macromolecule #4: Hemagglutinin HA2 chain
| Macromolecule | Name: Hemagglutinin HA2 chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Influenza A virus Influenza A virus |
| Molecular weight | Theoretical: 26.988895 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: GLFGAIAGFI EGGWTGMVDG WYGYHHQNEQ GSGYAADQKS TQNAINGITN KVNSVIEKMN TQFTAVGKEF NKLERRMENL NKKVDDGFI DIWTYNAELL VLLENERTLD FHDSNVKNLY EKVKSQLKNN AKEIGNGCFE FYHKCNDECM ESVKNGTYDY P KYSEESKL ...String: GLFGAIAGFI EGGWTGMVDG WYGYHHQNEQ GSGYAADQKS TQNAINGITN KVNSVIEKMN TQFTAVGKEF NKLERRMENL NKKVDDGFI DIWTYNAELL VLLENERTLD FHDSNVKNLY EKVKSQLKNN AKEIGNGCFE FYHKCNDECM ESVKNGTYDY P KYSEESKL NREKIDSGGG GLNDIFEAQK IEWHERLVPR GSPGSGYIPE APRDGQAYVR KDGEWVLLST FLGHHHHHH UniProtKB: Hemagglutinin |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 9 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.6 mg/mL |
|---|---|
| Buffer | pH: 7.6 / Details: Tris-buffered saline |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 25 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.8 µm / Nominal magnification: 29000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller








 Z
Z Y
Y X
X