+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Axle-less Bacillus sp. PS3 F1 ATPase mutant | |||||||||
 Map data Map data | Sharpened axle-less Bacillus PS3 ATPase map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | ATPase / ATP synthase / HYDROLASE | |||||||||
| Function / homology |  Function and homology information Function and homology informationproton motive force-driven plasma membrane ATP synthesis / proton-transporting ATP synthase complex, catalytic core F(1) / H+-transporting two-sector ATPase / proton-transporting ATPase activity, rotational mechanism / proton-transporting ATP synthase activity, rotational mechanism / ADP binding / ATP hydrolysis activity / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  | |||||||||
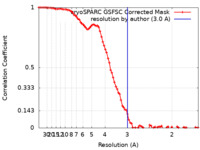
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Furlong EJ / Zeng YC / Brown SHJ / Sobti M / Stewart AG | |||||||||
| Funding support |  Australia, 1 items Australia, 1 items
| |||||||||
 Citation Citation |  Journal: Biochim Biophys Acta Bioenerg / Year: 2024 Journal: Biochim Biophys Acta Bioenerg / Year: 2024Title: The molecular structure of an axle-less F-ATPase. Authors: Emily J Furlong / Ian-Blaine P Reininger-Chatzigiannakis / Yi C Zeng / Simon H J Brown / Meghna Sobti / Alastair G Stewart /  Abstract: FF ATP synthase is a molecular rotary motor that can generate ATP using a transmembrane proton motive force. Isolated F-ATPase catalytic cores can hydrolyse ATP, passing through a series of ...FF ATP synthase is a molecular rotary motor that can generate ATP using a transmembrane proton motive force. Isolated F-ATPase catalytic cores can hydrolyse ATP, passing through a series of conformational states involving rotation of the central γ rotor subunit and the opening and closing of the catalytic β subunits. Cooperativity in F-ATPase has long thought to be conferred through the γ subunit, with three key interaction sites between the γ and β subunits being identified. Single molecule studies have demonstrated that the F complexes lacking the γ axle still "rotate" and hydrolyse ATP, but with less efficiency. We solved the cryogenic electron microscopy structure of an axle-less Bacillus sp. PS3 F-ATPase. The unexpected binding-dwell conformation of the structure in combination with the observed lack of interactions between the axle-less γ and the open β subunit suggests that the complete γ subunit is important for coordinating efficient ATP binding of F-ATPase. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41811.map.gz emd_41811.map.gz | 56.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41811-v30.xml emd-41811-v30.xml emd-41811.xml emd-41811.xml | 22.9 KB 22.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41811_fsc.xml emd_41811_fsc.xml | 8.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_41811.png emd_41811.png | 57.1 KB | ||
| Masks |  emd_41811_msk_1.map emd_41811_msk_1.map | 59.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-41811.cif.gz emd-41811.cif.gz | 7 KB | ||
| Others |  emd_41811_additional_1.map.gz emd_41811_additional_1.map.gz emd_41811_half_map_1.map.gz emd_41811_half_map_1.map.gz emd_41811_half_map_2.map.gz emd_41811_half_map_2.map.gz | 29.9 MB 55.4 MB 55.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41811 http://ftp.pdbj.org/pub/emdb/structures/EMD-41811 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41811 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41811 | HTTPS FTP |
-Validation report
| Summary document |  emd_41811_validation.pdf.gz emd_41811_validation.pdf.gz | 1.3 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41811_full_validation.pdf.gz emd_41811_full_validation.pdf.gz | 1.3 MB | Display | |
| Data in XML |  emd_41811_validation.xml.gz emd_41811_validation.xml.gz | 15.9 KB | Display | |
| Data in CIF |  emd_41811_validation.cif.gz emd_41811_validation.cif.gz | 20.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41811 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41811 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41811 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41811 | HTTPS FTP |
-Related structure data
| Related structure data |  8u1hMC  9avjC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41811.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41811.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened axle-less Bacillus PS3 ATPase map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.82 Å | ||||||||||||||||||||||||||||||||||||
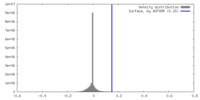
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_41811_msk_1.map emd_41811_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
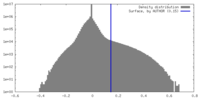
| Density Histograms |
- Sample components
Sample components
-Entire : Bacillus PS3 F1 ATPase complex, with truncated gamma subunit
| Entire | Name: Bacillus PS3 F1 ATPase complex, with truncated gamma subunit |
|---|---|
| Components |
|
-Supramolecule #1: Bacillus PS3 F1 ATPase complex, with truncated gamma subunit
| Supramolecule | Name: Bacillus PS3 F1 ATPase complex, with truncated gamma subunit type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 Details: The first 4 and last 25 amino acids have been deleted from the gamma subunit |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 354 KDa |
-Macromolecule #1: ATP synthase subunit alpha
| Macromolecule | Name: ATP synthase subunit alpha / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 54.908676 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSIRAEEISA LIKQQIENYE SQIQVSDVGT VIQVGDGIAR AHGLDNVMSG ELVEFANGVM GMALNLEENN VGIVILGPYT GIKEGDEVR RTGRIMEVPV GEALIGRVVN PLGQPVDGLG PVETTETRPI ESRAPGVMDR RSVHEPLQTG IKAIDALVPI G RGQRELII ...String: MSIRAEEISA LIKQQIENYE SQIQVSDVGT VIQVGDGIAR AHGLDNVMSG ELVEFANGVM GMALNLEENN VGIVILGPYT GIKEGDEVR RTGRIMEVPV GEALIGRVVN PLGQPVDGLG PVETTETRPI ESRAPGVMDR RSVHEPLQTG IKAIDALVPI G RGQRELII GDRQTGKTSV AIDTIINQKD QNMISIYVAI GQKESTVRTV VETLRKHGAL DYTIVVTASA SQPAPLLFLA PY AGVAMGE YFMYKGKHVL VVYDDLSKQA AAYRELSLLL RRPPGREAYP GDIFYLHSRL LERAAKLSDA KGGGSLTALP FVE TQAGDI SAYIPTNVIS ITDGQIFLQS DLFFSGVRPA INAGLSVSRV GGAAQIKAMK KVAGTLRLDL AAYRELEAFA QFGS DLDKA TQAKLARGAR TVEVLKQDLH QPIPVEKQVL IIYALTRGFL DDIPVEDVRR FEKEFYLFLD QNGQHLLEHI RTTKD LPNE DDLNKAIEAF KKTFVVSQ UniProtKB: ATP synthase subunit alpha |
-Macromolecule #2: ATP synthase subunit beta
| Macromolecule | Name: ATP synthase subunit beta / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 53.424625 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHHHH HMTRGRVIQV MGPVVDVKFE NGHLPAIYNA LKIQHKARNE NEVDIDLTLE VALHLGDDTV RTIAMASTDG LIRGMEVID TGAPISVPVG EVTLGRVFNV LGEPIDLEGD IPADARRDPI HRPAPKFEEL ATEVEILETG IKVVDLLAPY I KGGKIGLF ...String: MHHHHHHHHH HMTRGRVIQV MGPVVDVKFE NGHLPAIYNA LKIQHKARNE NEVDIDLTLE VALHLGDDTV RTIAMASTDG LIRGMEVID TGAPISVPVG EVTLGRVFNV LGEPIDLEGD IPADARRDPI HRPAPKFEEL ATEVEILETG IKVVDLLAPY I KGGKIGLF GGAGVGKTVL IQELIHNIAQ EHGGISVFAG VGERTREGND LYHEMKDSGV ISKTAMVFGQ MNEPPGARMR VA LTGLTMA EYFRDEQGQD VLLFIDNIFR FTQAGSEVSA LLGRMPSAVG YQPTLATEMG QLQERITSTA KGSITSIQAI YVP ADDYTD PAPATTFSHL DATTNLERKL AEMGIYPAVD PLASTSRALA PEIVGEEHYQ VARKVQQTLQ RYKELQDIIA ILGM DELSD EDKLVVHRAR RIQFFLSQNF HVAEQFTGQP GSYVPVKETV RGFKEILEGK YDHLPEDAFR LVGRIEEVVE KAKAM GVEV UniProtKB: ATP synthase subunit beta |
-Macromolecule #3: ATP synthase gamma chain
| Macromolecule | Name: ATP synthase gamma chain / type: protein_or_peptide / ID: 3 Details: 4 residues deleted from N-term, 25 residues deleted from C-term, S109C, I212C, Strep-tag II insertion at N197 Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 29.677084 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDIKTRINAT KKTSQITKAM EMVSTSKLNR AEQNAKSFVP YMEKIQEVVA NVALGAGGAS HPMLVSRPVK KTGYLVITSD RGLAGAYNS NVLRLVYQTI QKRHACPDEY AIIVIGRVGL SFFRKRNMPV ILDITRLPDQ PSFADIKEIA RKTVGLFADG T FDELYMYY ...String: MDIKTRINAT KKTSQITKAM EMVSTSKLNR AEQNAKSFVP YMEKIQEVVA NVALGAGGAS HPMLVSRPVK KTGYLVITSD RGLAGAYNS NVLRLVYQTI QKRHACPDEY AIIVIGRVGL SFFRKRNMPV ILDITRLPDQ PSFADIKEIA RKTVGLFADG T FDELYMYY NHYVSAIQQE VTERKLLPLT DLAENWSHPQ FEKQRTVYEF EPSQEECLDV LLPQYAESLI YGALLDAKAS EH AARMTAM KNATDNANEL IRTLTL UniProtKB: ATP synthase gamma chain |
-Macromolecule #4: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 4 / Number of copies: 4 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Macromolecule #5: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 5 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #6: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 6 / Number of copies: 1 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #7: PHOSPHATE ION
| Macromolecule | Name: PHOSPHATE ION / type: ligand / ID: 7 / Number of copies: 2 / Formula: PO4 |
|---|---|
| Molecular weight | Theoretical: 94.971 Da |
| Chemical component information |  ChemComp-PO4: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 6 mg/mL |
|---|---|
| Buffer | pH: 7 Details: 100 mM Potassium phosphate pH 7, 2 mM EDTA 1 mM AMPPNP was added 20 min prior to freezing |
| Grid | Model: UltrAuFoil R1.2/1.3 / Material: GOLD / Mesh: 300 / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. / Pretreatment - Atmosphere: AIR Details: Grid was glow discharged using a PELCO easiGlow set to 15 mA |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 293 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 59.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 0.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | ModelAngelo was used to build an initial model into the map |
|---|---|
| Refinement | Space: REAL / Protocol: AB INITIO MODEL |
| Output model |  PDB-8u1h: |
 Movie
Movie Controller
Controller








 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)