[English] 日本語
 Yorodumi
Yorodumi- EMDB-41498: HIV-1 BG505 Env SOSIP in complex with bovine Fab Bess4 and non-hu... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | HIV-1 BG505 Env SOSIP in complex with bovine Fab Bess4 and non-human primate Fab RM20A3 | |||||||||
 Map data Map data | sharpened map | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SOSIP / apex / broadly neutralizing antibody / Env / HIV-1 / VIRAL PROTEIN / VIRAL PROTEIN-Immune System complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationpositive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / : / viral envelope / virion attachment to host cell ...positive regulation of plasma membrane raft polarization / positive regulation of receptor clustering / host cell endosome membrane / clathrin-dependent endocytosis of virus by host cell / viral protein processing / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / : / viral envelope / virion attachment to host cell / host cell plasma membrane / virion membrane / structural molecule activity / identical protein binding / membrane Similarity search - Function | |||||||||
| Biological species |   Human immunodeficiency virus 1 / Human immunodeficiency virus 1 /   | |||||||||
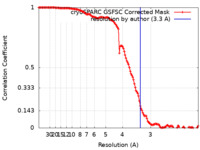
| Method | single particle reconstruction / cryo EM / Resolution: 3.3 Å | |||||||||
 Authors Authors | Ozorowski G / Lee WH / Ward AB | |||||||||
| Funding support |  United States, 1 items United States, 1 items
| |||||||||
 Citation Citation |  Journal: PLoS Pathog / Year: 2024 Journal: PLoS Pathog / Year: 2024Title: Immunization of cows with HIV envelope trimers generates broadly neutralizing antibodies to the V2-apex from the ultralong CDRH3 repertoire. Authors: Pilar X Altman / Gabriel Ozorowski / Robyn L Stanfield / Jeremy Haakenson / Michael Appel / Mara Parren / Wen-Hsin Lee / Huldah Sang / Jordan Woehl / Karen Saye-Francisco / Leigh M Sewall / ...Authors: Pilar X Altman / Gabriel Ozorowski / Robyn L Stanfield / Jeremy Haakenson / Michael Appel / Mara Parren / Wen-Hsin Lee / Huldah Sang / Jordan Woehl / Karen Saye-Francisco / Leigh M Sewall / Collin Joyce / Ge Song / Katelyn Porter / Elise Landais / Raiees Andrabi / Ian A Wilson / Andrew B Ward / Waithaka Mwangi / Vaughn V Smider / Dennis R Burton / Devin Sok /  Abstract: The generation of broadly neutralizing antibodies (bnAbs) to conserved epitopes on HIV Envelope (Env) is one of the cornerstones of HIV vaccine research. The animal models commonly used for HIV do ...The generation of broadly neutralizing antibodies (bnAbs) to conserved epitopes on HIV Envelope (Env) is one of the cornerstones of HIV vaccine research. The animal models commonly used for HIV do not reliably produce a potent broadly neutralizing serum antibody response, with the exception of cows. Cows have previously produced a CD4 binding site response by homologous prime and boosting with a native-like Env trimer. In small animal models, other engineered immunogens were shown to focus antibody responses to the bnAb V2-apex region of Env. Here, we immunized two groups of cows (n = 4) with two regimens of V2-apex focusing Env immunogens to investigate whether antibody responses could be generated to the V2-apex on Env. Group 1 was immunized with chimpanzee simian immunodeficiency virus (SIV)-Env trimer that shares its V2-apex with HIV, followed by immunization with C108, a V2-apex focusing immunogen, and finally boosted with a cross-clade native-like trimer cocktail. Group 2 was immunized with HIV C108 Env trimer followed by the same HIV trimer cocktail as Group 1. Longitudinal serum analysis showed that one cow in each group developed serum neutralizing antibody responses to the V2-apex. Eight and 11 bnAbs were isolated from Group 1 and Group 2 cows, respectively, and showed moderate breadth and potency. Potent and broad responses in this study developed much later than previous cow immunizations that elicited CD4bs bnAbs responses and required several different immunogens. All isolated bnAbs were derived from the ultralong CDRH3 repertoire. The finding that cow antibodies can target more than one broadly neutralizing epitope on the HIV surface reveals the generality of elongated structures for the recognition of highly glycosylated proteins. The exclusive isolation of ultralong CDRH3 bnAbs, despite only comprising a small percent of the cow repertoire, suggests these antibodies outcompete the long and short CDRH3 antibodies during the bnAb response. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_41498.map.gz emd_41498.map.gz | 204.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-41498-v30.xml emd-41498-v30.xml emd-41498.xml emd-41498.xml | 21.3 KB 21.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_41498_fsc.xml emd_41498_fsc.xml | 14.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_41498.png emd_41498.png | 116.1 KB | ||
| Masks |  emd_41498_msk_1.map emd_41498_msk_1.map | 216 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-41498.cif.gz emd-41498.cif.gz | 7.1 KB | ||
| Others |  emd_41498_half_map_1.map.gz emd_41498_half_map_1.map.gz emd_41498_half_map_2.map.gz emd_41498_half_map_2.map.gz | 200.7 MB 200.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41498 http://ftp.pdbj.org/pub/emdb/structures/EMD-41498 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41498 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41498 | HTTPS FTP |
-Validation report
| Summary document |  emd_41498_validation.pdf.gz emd_41498_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_41498_full_validation.pdf.gz emd_41498_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_41498_validation.xml.gz emd_41498_validation.xml.gz | 20.9 KB | Display | |
| Data in CIF |  emd_41498_validation.cif.gz emd_41498_validation.cif.gz | 26.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41498 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41498 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41498 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-41498 | HTTPS FTP |
-Related structure data
| Related structure data |  8tq1MC  8v4iC  8vbjC  8vbkC  8vblC  8vbmC  8vbnC  8vboC  8vbpC  8vbqC  8vbrC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_41498.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_41498.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | sharpened map | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||
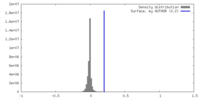
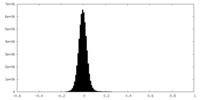
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_41498_msk_1.map emd_41498_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
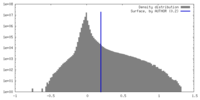
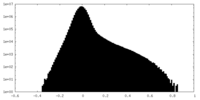
| Density Histograms |
-Half map: half map B
| File | emd_41498_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map B | ||||||||||||
| Projections & Slices |
| ||||||||||||
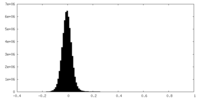
| Density Histograms |
-Half map: half map A
| File | emd_41498_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map A | ||||||||||||
| Projections & Slices |
| ||||||||||||
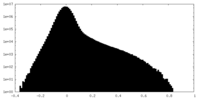
| Density Histograms |
- Sample components
Sample components
-Entire : HIV-1 BG505 SOSIP Env in complex with bovine Bess4 Fab and NHP RM...
| Entire | Name: HIV-1 BG505 SOSIP Env in complex with bovine Bess4 Fab and NHP RM20A3 Fab |
|---|---|
| Components |
|
-Supramolecule #1: HIV-1 BG505 SOSIP Env in complex with bovine Bess4 Fab and NHP RM...
| Supramolecule | Name: HIV-1 BG505 SOSIP Env in complex with bovine Bess4 Fab and NHP RM20A3 Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
-Macromolecule #1: HIV-1 Envelope Glycoprotein BG505 SOSIP.664 gp120
| Macromolecule | Name: HIV-1 Envelope Glycoprotein BG505 SOSIP.664 gp120 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 57.945977 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARAENLW VTVYYGVPVW KDAETTLFCA SDAKAYETEK HNVWATHACV PTDPNPQEI HLENVTEEFN MWKNNMVEQM HTDIISLWDQ SLKPCVKLTP LCVTLQCTNV TNNITDDMRG ELKNCSFNMT T ELRDKKQK ...String: MDAMKRGLCC VLLLCGAVFV SPSQEIHARF RRGARAENLW VTVYYGVPVW KDAETTLFCA SDAKAYETEK HNVWATHACV PTDPNPQEI HLENVTEEFN MWKNNMVEQM HTDIISLWDQ SLKPCVKLTP LCVTLQCTNV TNNITDDMRG ELKNCSFNMT T ELRDKKQK VYSLFYRLDV VQINENQGNR SNNSNKEYRL INCNTSAITQ ACPKVSFEPI PIHYCAPAGF AILKCKDKKF NG TGPCPSV STVQCTHGIK PVVSTQLLLN GSLAEEEVMI RSENITNNAK NILVQFNTPV QINCTRPNNN TRKSIRIGPG QAF YATGDI IGDIRQAHCN VSKATWNETL GKVVKQLRKH FGNNTIIRFA NSSGGDLEVT THSFNCGGEF FYCNTSGLFN STWI SNTSV QGSNSTGSND SITLPCRIKQ IINMWQRIGQ AMYAPPIQGV IRCVSNITGL ILTRDGGSTN STTETFRPGG GDMRD NWRS ELYKYKVVKI EPLGVAPTRC KRRVVGRRRR RR UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #2: Transmembrane protein gp41
| Macromolecule | Name: Transmembrane protein gp41 / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Molecular weight | Theoretical: 17.146482 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: AVGIGAVFLG FLGAAGSTMG AASMTLTVQA RNLLSGIVQQ QSNLLRAPEA QQHLLKLTVW GIKQLQARVL AVERYLRDQQ LLGIWGCSG KLICCTNVPW NSSWSNRNLS EIWDNMTWLQ WDKEISNYTQ IIYGLLEESQ NQQEKNEQDL LALD UniProtKB: Envelope glycoprotein gp160 |
-Macromolecule #3: NHP RM20A3 Fab heavy chain
| Macromolecule | Name: NHP RM20A3 Fab heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.511111 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: EVQLVETGGG LVQPGGSLKL SCRASGYTFS SFAMSWVRQA PGKGLEWVSL INDRGGLTFY VDSVKGRFTI SRDNSKNTLS LQMHSLRDG DTAVYYCATG GMSSALQSSK YYFDFWGQGA LVTVSS |
-Macromolecule #4: NHP RM20A3 Fab light chain
| Macromolecule | Name: NHP RM20A3 Fab light chain / type: protein_or_peptide / ID: 4 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.5088 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: ALTQPPSVSG SPGQSVTISC TGTSSDIGSY NYVSWYQQHP GKAPKLMIYD VTQRPSGVSD RFSGSKSGNT ASLTISGLQA DDEADYYCS AYAGRQTFYI FGGGTRLTVL GQPKASPTVT LFPPSSEEL |
-Macromolecule #5: Bovine Bess4 Fab heavy chain
| Macromolecule | Name: Bovine Bess4 Fab heavy chain / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 17.435293 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: KVQLRESGPS LVKPSQTLSL TCTASGLTLS DKAVGWVRQA PGKALEWLGS IDTSGNTGYN PGLKSRLTIT KDSSKSQVSL SVSSVTTED SATYYCTTVH QQTRNKVKSC PSGEDCGIGC CYHGCSATDY GCWDGSSYAP YSYTYTYELH VDTWGQGLLV T VSS |
-Macromolecule #9: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 9 / Number of copies: 45 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 2.5 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 55.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.7 µm / Nominal defocus min: 0.7000000000000001 µm / Nominal magnification: 130000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)