+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structure of the TUG-891 bound FFA4-Gq complex | |||||||||
 マップデータ マップデータ | TUG-891-FFA4-miniGq | |||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | GPCR / Complex / agonist / MEMBRANE PROTEIN | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報negative regulation of somatostatin secretion / positive regulation of glucagon secretion / ghrelin secretion / Free fatty acid receptors / regulation of D-glucose transmembrane transport / taste receptor activity / hormone secretion / arrestin family protein binding / negative regulation of cytokine production / negative regulation of interleukin-1 beta production ...negative regulation of somatostatin secretion / positive regulation of glucagon secretion / ghrelin secretion / Free fatty acid receptors / regulation of D-glucose transmembrane transport / taste receptor activity / hormone secretion / arrestin family protein binding / negative regulation of cytokine production / negative regulation of interleukin-1 beta production / ciliary membrane / white fat cell differentiation / endocytic vesicle / positive regulation of osteoblast differentiation / positive regulation of brown fat cell differentiation / brown fat cell differentiation / fatty acid binding / G protein-coupled receptor activity / negative regulation of inflammatory response / Olfactory Signaling Pathway / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Glucagon-type ligand receptors / Sensory perception of sweet, bitter, and umami (glutamate) taste / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / Inactivation, recovery and regulation of the phototransduction cascade / extracellular vesicle / sensory perception of taste / positive regulation of cold-induced thermogenesis / Thrombin signalling through proteinase activated receptors (PARs) / signaling receptor complex adaptor activity / retina development in camera-type eye / positive regulation of cytosolic calcium ion concentration / GTPase binding / Ca2+ pathway / fibroblast proliferation / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / Ras protein signal transduction / Extra-nuclear estrogen signaling / positive regulation of ERK1 and ERK2 cascade / cell population proliferation / endosome membrane / cilium / ciliary basal body / G protein-coupled receptor signaling pathway / inflammatory response / intracellular membrane-bounded organelle / lysosomal membrane / GTPase activity / synapse / negative regulation of apoptotic process / protein-containing complex binding / signal transduction / extracellular exosome / membrane / plasma membrane / cytoplasm / cytosol 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) / Homo sapiens (ヒト) /  | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.06 Å | |||||||||
 データ登録者 データ登録者 | Zhang X / Tikhonova I / Milligan G / Zhang C | |||||||||
| 資金援助 |  米国, 1件 米国, 1件
| |||||||||
 引用 引用 |  ジャーナル: Sci Adv / 年: 2024 ジャーナル: Sci Adv / 年: 2024タイトル: Structural basis for the ligand recognition and signaling of free fatty acid receptors. 著者: Xuan Zhang / Abdul-Akim Guseinov / Laura Jenkins / Kunpeng Li / Irina G Tikhonova / Graeme Milligan / Cheng Zhang /   要旨: Free fatty acid receptors 1 to 4 (FFA1 to FFA4) are class A G protein-coupled receptors (GPCRs). FFA1 to FFA3 share substantial sequence similarity, whereas FFA4 is unrelated. However, FFA1 and FFA4 ...Free fatty acid receptors 1 to 4 (FFA1 to FFA4) are class A G protein-coupled receptors (GPCRs). FFA1 to FFA3 share substantial sequence similarity, whereas FFA4 is unrelated. However, FFA1 and FFA4 are activated by long-chain fatty acids, while FFA2 and FFA3 respond to short-chain fatty acids generated by intestinal microbiota. FFA1, FFA2, and FFA4 are potential drug targets for metabolic and inflammatory conditions. Here, we determined the active structures of FFA1 and FFA4 bound to docosahexaenoic acid, FFA4 bound to the synthetic agonist TUG-891, and butyrate-bound FFA2, each complexed with an engineered heterotrimeric G protein (miniG), by cryo-electron microscopy. Together with computational simulations and mutagenesis studies, we elucidated the similarities and differences in the binding modes of fatty acid ligands to their respective GPCRs. Our findings unveiled distinct mechanisms of receptor activation and G protein coupling. We anticipate that these outcomes will facilitate structure-based drug development and underpin future research on this group of GPCRs. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_41007.map.gz emd_41007.map.gz | 59.7 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-41007-v30.xml emd-41007-v30.xml emd-41007.xml emd-41007.xml | 22 KB 22 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_41007.png emd_41007.png | 30.3 KB | ||
| Filedesc metadata |  emd-41007.cif.gz emd-41007.cif.gz | 7.1 KB | ||
| その他 |  emd_41007_half_map_1.map.gz emd_41007_half_map_1.map.gz emd_41007_half_map_2.map.gz emd_41007_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-41007 http://ftp.pdbj.org/pub/emdb/structures/EMD-41007 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41007 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-41007 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_41007.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_41007.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | TUG-891-FFA4-miniGq | ||||||||||||||||||||||||||||||||||||
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.828 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-ハーフマップ: TUG-891-FFA4-miniGq-half A
| ファイル | emd_41007_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | TUG-891-FFA4-miniGq-half_A | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: TUG-891-FFA4-miniGq-half B
| ファイル | emd_41007_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | TUG-891-FFA4-miniGq-half_B | ||||||||||||
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : TUG-891-FFA4-miniGq complex
| 全体 | 名称: TUG-891-FFA4-miniGq complex |
|---|---|
| 要素 |
|
-超分子 #1: TUG-891-FFA4-miniGq complex
| 超分子 | 名称: TUG-891-FFA4-miniGq complex / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1-#5 |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-分子 #1: Free fatty acid receptor 4
| 分子 | 名称: Free fatty acid receptor 4 / タイプ: protein_or_peptide / ID: 1 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 33.517266 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: RTRFPFFSDV KGDHRLVLAA VETTVLVLIF AVSLLGNVCA LVLVARRRRR GATACLVLNL FCADLLFISA IPLVLAVRWT EAWLLGPVA CHLLFYVMTL SGSVTILTLA AVSLERMVCI VHLQRGPGRR ARAVLLALIW GYSAVAALPL CVFFRVVPQR I SICTLIWP ...文字列: RTRFPFFSDV KGDHRLVLAA VETTVLVLIF AVSLLGNVCA LVLVARRRRR GATACLVLNL FCADLLFISA IPLVLAVRWT EAWLLGPVA CHLLFYVMTL SGSVTILTLA AVSLERMVCI VHLQRGPGRR ARAVLLALIW GYSAVAALPL CVFFRVVPQR I SICTLIWP TIPGEISWDV SFVTLNFLVP GLVIVISYSK ILQITKASRK RLTVSLAYSE SHQIRVSQQD FRLFRTLFLL MV SFFIMWS PIIITILLIL IQNFKQDLVI WPSLFFWVVA FTFANSALNP ILYNMTLCRN UniProtKB: Free fatty acid receptor 4 |
-分子 #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| 分子 | 名称: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 タイプ: protein_or_peptide / ID: 2 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 37.516941 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: SELDQLRQEA EQLKNQIRDA RKACADATLS QITNNIDPVG RIQMRTRRTL RGHLAKIYAM HWGTDSRLLV SASQDGKLII WDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI CSIYNLKTRE GNVRVSRELA GHTGYLSCCR FLDDNQIVTS S GDTTCALW ...文字列: SELDQLRQEA EQLKNQIRDA RKACADATLS QITNNIDPVG RIQMRTRRTL RGHLAKIYAM HWGTDSRLLV SASQDGKLII WDSYTTNKV HAIPLRSSWV MTCAYAPSGN YVACGGLDNI CSIYNLKTRE GNVRVSRELA GHTGYLSCCR FLDDNQIVTS S GDTTCALW DIETGQQTTT FTGHTGDVMS LSLAPDTRLF VSGACDASAK LWDVREGMCR QTFTGHESDI NAICFFPNGN AF ATGSDDA TCRLFDLRAD QELMTYSHDN IICGITSVSF SKSGRLLLAG YDDFNCNVWD ALKADRAGVL AGHDNRVSCL GVT DDGMAV ATGSWDSFLK IWNGSS UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-分子 #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| 分子 | 名称: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 タイプ: protein_or_peptide / ID: 3 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 6.261229 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: TASIAQARKL VEQLKMEANI DRIKVSKAAA DLMAYCEAHA KEDPLLTPVP ASENPFR UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-分子 #4: Guanine nucleotide-binding protein G(q)
| 分子 | 名称: Guanine nucleotide-binding protein G(q) / タイプ: protein_or_peptide / ID: 4 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 26.670275 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: VSAEDKAAAE RSKMIDKNLR EDGEKARRTL RLLLLGADNS GKSTIVKQMT SGIFETKFQV DKVNFHMFDV GGQRDERRKW IQCFNDVTA IIFVVDSSDY NRLQEALNDF KSIWNNRWLR TISVILFLNK QDLLAEKVLA GKSKIEDYFP EFARYTTPED A TPEPGEDP ...文字列: VSAEDKAAAE RSKMIDKNLR EDGEKARRTL RLLLLGADNS GKSTIVKQMT SGIFETKFQV DKVNFHMFDV GGQRDERRKW IQCFNDVTA IIFVVDSSDY NRLQEALNDF KSIWNNRWLR TISVILFLNK QDLLAEKVLA GKSKIEDYFP EFARYTTPED A TPEPGEDP RVTRAKYFIR KEFVDISTAS GDGRHICYPH FTCAVDTENA RRIFNDCKDI ILQMNLREYN LV |
-分子 #5: scFv16
| 分子 | 名称: scFv16 / タイプ: protein_or_peptide / ID: 5 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 28.634797 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS AGGGGSGGGG SGGGGSADIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...文字列: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS AGGGGSGGGG SGGGGSADIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL LEENLYFQGA SHHHHHHHH |
-分子 #6: 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}prop...
| 分子 | 名称: 3-{4-[(4-fluoro-4'-methyl[1,1'-biphenyl]-2-yl)methoxy]phenyl}propanoic acid タイプ: ligand / ID: 6 / コピー数: 1 / 式: YN9 |
|---|---|
| 分子量 | 理論値: 364.409 Da |
| Chemical component information | 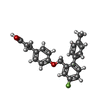 ChemComp-YN9: |
-分子 #7: (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-...
| 分子 | 名称: (2R)-1-{[(R)-hydroxy{[(1R,2R,3R,4R,5S,6R)-2,3,5,6-tetrahydroxy-4-(phosphonooxy)cyclohexyl]oxy}phosphoryl]oxy}-3-(octadecanoyloxy)propan-2-yl (5Z,8Z,11Z,14Z)-icosa-5,8,11,14-tetraenoate タイプ: ligand / ID: 7 / コピー数: 1 / 式: 2Y5 |
|---|---|
| 分子量 | 理論値: 967.108 Da |
| Chemical component information |  ChemComp-2Y5: |
-分子 #8: PALMITIC ACID
| 分子 | 名称: PALMITIC ACID / タイプ: ligand / ID: 8 / コピー数: 1 / 式: PLM |
|---|---|
| 分子量 | 理論値: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.5 |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 BIOQUANTUM (6k x 4k) 平均電子線量: 55.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 1.8 µm / 最小 デフォーカス(公称値): 1.0 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
 ムービー
ムービー コントローラー
コントローラー

































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)