+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Ciliary cap complex from Tetrahymena thermophila | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cilia / Axoneme / central pair / microtubules / STRUCTURAL PROTEIN | |||||||||
| Biological species |  | |||||||||
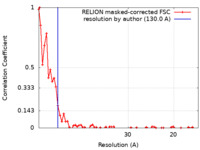
| Method | subtomogram averaging / cryo EM / Resolution: 130.0 Å | |||||||||
 Authors Authors | Legal T / Bui KH | |||||||||
| Funding support |  Canada, 1 items Canada, 1 items
| |||||||||
 Citation Citation |  Journal: J Cell Biol / Year: 2023 Journal: J Cell Biol / Year: 2023Title: CEP104/FAP256 and associated cap complex maintain stability of the ciliary tip. Authors: Thibault Legal / Mireya Parra / Maxwell Tong / Corbin S Black / Ewa Joachimiak / Melissa Valente-Paterno / Karl Lechtreck / Jacek Gaertig / Khanh Huy Bui /    Abstract: Cilia are essential organelles that protrude from the cell body. Cilia are made of a microtubule-based structure called the axoneme. In most types of cilia, the ciliary tip is distinct from the rest ...Cilia are essential organelles that protrude from the cell body. Cilia are made of a microtubule-based structure called the axoneme. In most types of cilia, the ciliary tip is distinct from the rest of the cilium. Here, we used cryo-electron tomography and subtomogram averaging to obtain the structure of the ciliary tip of the ciliate Tetrahymena thermophila. We show that the microtubules at the tip are highly crosslinked with each other and stabilized by luminal proteins, plugs, and cap proteins at the plus ends. In the tip region, the central pair lacks typical projections and twists significantly. By analyzing cells lacking a ciliary tip-enriched protein CEP104/FAP256 by cryo-electron tomography and proteomics, we discovered candidates for the central pair cap complex and explained the potential functions of CEP104/FAP256. These data provide new insights into the function of the ciliary tip and the mechanisms of ciliary assembly and length regulation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40892.map.gz emd_40892.map.gz | 48.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40892-v30.xml emd-40892-v30.xml emd-40892.xml emd-40892.xml | 12 KB 12 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40892_fsc.xml emd_40892_fsc.xml | 6.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_40892.png emd_40892.png | 83.5 KB | ||
| Filedesc metadata |  emd-40892.cif.gz emd-40892.cif.gz | 3.8 KB | ||
| Others |  emd_40892_half_map_1.map.gz emd_40892_half_map_1.map.gz emd_40892_half_map_2.map.gz emd_40892_half_map_2.map.gz | 13.8 MB 13.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40892 http://ftp.pdbj.org/pub/emdb/structures/EMD-40892 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40892 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40892 | HTTPS FTP |
-Validation report
| Summary document |  emd_40892_validation.pdf.gz emd_40892_validation.pdf.gz | 983 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40892_full_validation.pdf.gz emd_40892_full_validation.pdf.gz | 982.5 KB | Display | |
| Data in XML |  emd_40892_validation.xml.gz emd_40892_validation.xml.gz | 12.4 KB | Display | |
| Data in CIF |  emd_40892_validation.cif.gz emd_40892_validation.cif.gz | 16.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40892 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40892 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40892 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40892 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_40892.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40892.map.gz / Format: CCP4 / Size: 52.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 8.48 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_40892_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
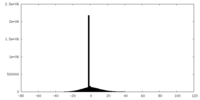
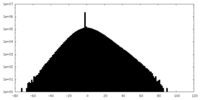
| Density Histograms |
-Half map: #2
| File | emd_40892_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
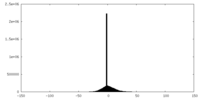
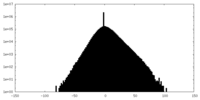
| Density Histograms |
- Sample components
Sample components
-Entire : Ciliary cap complex from Tetrahymena thermphila
| Entire | Name: Ciliary cap complex from Tetrahymena thermphila |
|---|---|
| Components |
|
-Supramolecule #1: Ciliary cap complex from Tetrahymena thermphila
| Supramolecule | Name: Ciliary cap complex from Tetrahymena thermphila / type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Concentration | 1.8 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 4.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 4.0 µm / Nominal defocus min: 2.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller

















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)