+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structure of the CBC-ALYREF complex | |||||||||
 マップデータ マップデータ | ||||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | mRNA nuclear export / RNA BINDING PROTEIN | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報positive regulation of RNA binding / snRNA export from nucleus / nuclear cap binding complex / histone mRNA metabolic process / RNA cap binding complex / mRNA metabolic process / positive regulation of RNA export from nucleus / positive regulation of mRNA 3'-end processing / cap-dependent translational initiation / Processing of Intronless Pre-mRNAs ...positive regulation of RNA binding / snRNA export from nucleus / nuclear cap binding complex / histone mRNA metabolic process / RNA cap binding complex / mRNA metabolic process / positive regulation of RNA export from nucleus / positive regulation of mRNA 3'-end processing / cap-dependent translational initiation / Processing of Intronless Pre-mRNAs / snRNA binding / RNA cap binding / alternative mRNA splicing, via spliceosome / miRNA-mediated post-transcriptional gene silencing / primary miRNA processing / regulation of mRNA processing / SLBP independent Processing of Histone Pre-mRNAs / SLBP Dependent Processing of Replication-Dependent Histone Pre-mRNAs / regulatory ncRNA-mediated post-transcriptional gene silencing / RNA 7-methylguanosine cap binding / Transport of the SLBP independent Mature mRNA / Transport of the SLBP Dependant Mature mRNA / mRNA 3'-end processing / Transport of Mature mRNA Derived from an Intronless Transcript / positive regulation of mRNA splicing, via spliceosome / mRNA 3'-end processing / mRNA cis splicing, via spliceosome / RNA catabolic process / Transport of Mature mRNA derived from an Intron-Containing Transcript / regulation of translational initiation / Abortive elongation of HIV-1 transcript in the absence of Tat / RNA Polymerase II Transcription Termination / nuclear-transcribed mRNA catabolic process, nonsense-mediated decay / FGFR2 alternative splicing / Signaling by FGFR2 IIIa TM / Formation of the Early Elongation Complex / Formation of the HIV-1 Early Elongation Complex / mRNA Capping / spliceosomal complex assembly / mRNA Splicing - Minor Pathway / Processing of Capped Intron-Containing Pre-mRNA / RNA polymerase II transcribes snRNA genes / Nonsense Mediated Decay (NMD) independent of the Exon Junction Complex (EJC) / 7-methylguanosine mRNA capping / Formation of HIV-1 elongation complex containing HIV-1 Tat / mRNA export from nucleus / Formation of HIV elongation complex in the absence of HIV Tat / Nonsense Mediated Decay (NMD) enhanced by the Exon Junction Complex (EJC) / Formation of RNA Pol II elongation complex / RNA Polymerase II Pre-transcription Events / mRNA Splicing - Major Pathway / RNA splicing / positive regulation of transcription elongation by RNA polymerase II / mRNA transcription by RNA polymerase II / mRNA splicing, via spliceosome / Regulation of expression of SLITs and ROBOs / snRNP Assembly / positive regulation of cell growth / molecular adaptor activity / defense response to virus / ribonucleoprotein complex / mRNA binding / mitochondrion / DNA binding / RNA binding / nucleoplasm / nucleus / cytosol / cytoplasm 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) / Homo sapiens (ヒト) /  | |||||||||
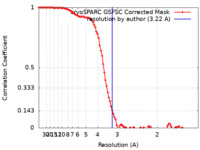
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.22 Å | |||||||||
 データ登録者 データ登録者 | Xie Y / Clarke BP / Ren Y | |||||||||
| 資金援助 |  米国, 1件 米国, 1件
| |||||||||
 引用 引用 |  ジャーナル: Elife / 年: 2024 ジャーナル: Elife / 年: 2024タイトル: Cryo-EM structure of the CBC-ALYREF complex. 著者: Bradley P Clarke / Alexia E Angelos / Menghan Mei / Pate S Hill / Yihu Xie / Yi Ren /  要旨: In eukaryotes, RNAs transcribed by RNA Pol II are modified at the 5' end with a 7-methylguanosine (mG) cap, which is recognized by the nuclear cap binding complex (CBC). The CBC plays multiple ...In eukaryotes, RNAs transcribed by RNA Pol II are modified at the 5' end with a 7-methylguanosine (mG) cap, which is recognized by the nuclear cap binding complex (CBC). The CBC plays multiple important roles in mRNA metabolism, including transcription, splicing, polyadenylation, and export. It promotes mRNA export through direct interaction with a key mRNA export factor, ALYREF, which in turn links the TRanscription and EXport (TREX) complex to the 5' end of mRNA. However, the molecular mechanism for CBC-mediated recruitment of the mRNA export machinery is not well understood. Here, we present the first structure of the CBC in complex with an mRNA export factor, ALYREF. The cryo-EM structure of CBC-ALYREF reveals that the RRM domain of ALYREF makes direct contact with both the NCBP1 and NCBP2 subunits of the CBC. Comparing CBC-ALYREF with other cellular complexes containing CBC and/or ALYREF components provides insights into the coordinated events during mRNA transcription, splicing, and export. #1:  ジャーナル: Elife / 年: 2024 ジャーナル: Elife / 年: 2024タイトル: Cryo-EM structure of the CBC-ALYREF complex 著者: Clarke BP / Angelos AE / Mei M / Hill PS / Xie Y / Ren Y | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_40739.map.gz emd_40739.map.gz | 45.5 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-40739-v30.xml emd-40739-v30.xml emd-40739.xml emd-40739.xml | 17.1 KB 17.1 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_40739_fsc.xml emd_40739_fsc.xml | 9.6 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_40739.png emd_40739.png | 81.7 KB | ||
| Filedesc metadata |  emd-40739.cif.gz emd-40739.cif.gz | 6 KB | ||
| その他 |  emd_40739_half_map_1.map.gz emd_40739_half_map_1.map.gz emd_40739_half_map_2.map.gz emd_40739_half_map_2.map.gz | 84.5 MB 84.5 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40739 http://ftp.pdbj.org/pub/emdb/structures/EMD-40739 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40739 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40739 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_40739_validation.pdf.gz emd_40739_validation.pdf.gz | 1.2 MB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_40739_full_validation.pdf.gz emd_40739_full_validation.pdf.gz | 1.2 MB | 表示 | |
| XML形式データ |  emd_40739_validation.xml.gz emd_40739_validation.xml.gz | 17.8 KB | 表示 | |
| CIF形式データ |  emd_40739_validation.cif.gz emd_40739_validation.cif.gz | 22.9 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40739 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40739 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40739 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40739 | HTTPS FTP |
-関連構造データ
| 関連構造データ |  8srrM M: このマップから作成された原子モデル C: 同じ文献を引用 ( |
|---|---|
| 類似構造データ | 類似検索 - 機能・相同性  F&H 検索 F&H 検索 |
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_40739.map.gz / 形式: CCP4 / 大きさ: 91.1 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_40739.map.gz / 形式: CCP4 / 大きさ: 91.1 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 0.732 Å | ||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-ハーフマップ: #2
| ファイル | emd_40739_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
-ハーフマップ: #1
| ファイル | emd_40739_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Cryo-EM structure of an mRNA export factor
| 全体 | 名称: Cryo-EM structure of an mRNA export factor |
|---|---|
| 要素 |
|
-超分子 #1: Cryo-EM structure of an mRNA export factor
| 超分子 | 名称: Cryo-EM structure of an mRNA export factor / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1-#3 |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-分子 #1: Nuclear cap-binding protein subunit 1
| 分子 | 名称: Nuclear cap-binding protein subunit 1 / タイプ: protein_or_peptide / ID: 1 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 91.960297 KDa |
| 組換発現 | 生物種:  Trichoplusia ni (イラクサキンウワバ) Trichoplusia ni (イラクサキンウワバ) |
| 配列 | 文字列: MSRRRHSDEN DGGQPHKRRK TSDANETEDH LESLICKVGE KSACSLESNL EGLAGVLEAD LPNYKSKILR LLCTVARLLP EKLTIYTTL VGLLNARNYN FGGEFVEAMI RQLKESLKAN NYNEAVYLVR FLSDLVNCHV IAAPSMVAMF ENFVSVTQEE D VPQVRRDW ...文字列: MSRRRHSDEN DGGQPHKRRK TSDANETEDH LESLICKVGE KSACSLESNL EGLAGVLEAD LPNYKSKILR LLCTVARLLP EKLTIYTTL VGLLNARNYN FGGEFVEAMI RQLKESLKAN NYNEAVYLVR FLSDLVNCHV IAAPSMVAMF ENFVSVTQEE D VPQVRRDW YVYAFLSSLP WVGKELYEKK DAEMDRIFAN TESYLKRRQK THVPMLQVWT ADKPHPQEEY LDCLWAQIQK LK KDRWQER HILRPYLAFD SILCEALQHN LPPFTPPPHT EDSVYPMPRV IFRMFDYTDD PEGPVMPGSH SVERFVIEEN LHC IIKSHW KERKTCAAQL VSYPGKNKIP LNYHIVEVIF AELFQLPAPP HIDVMYTTLL IELCKLQPGS LPQVLAQATE MLYM RLDTM NTTCVDRFIN WFSHHLSNFQ FRWSWEDWSD CLSQDPESPK PKFVREVLEK CMRLSYHQRI LDIVPPTFSA LCPAN PTCI YKYGDESSNS LPGHSVALCL AVAFKSKATN DEIFSILKDV PNPNQDDDDD EGFSFNPLKI EVFVQTLLHL AAKSFS HSF SALAKFHEVF KTLAESDEGK LHVLRVMFEV WRNHPQMIAV LVDKMIRTQI VDCAAVANWI FSSELSRDFT RLFVWEI LH STIRKMNKHV LKIQKELEEA KEKLARQHKR RSDDDDRSSD RKDGVLEEQI ERLQEKVESA QSEQKNLFLV IFQRFIMI L TEHLVRCETD GTSVLTPWYK NCIERLQQIF LQHHQIIQQY MVTLENLLFT AELDPHILAV FQQFCALQA UniProtKB: Nuclear cap-binding protein subunit 1 |
-分子 #2: Nuclear cap-binding protein subunit 2
| 分子 | 名称: Nuclear cap-binding protein subunit 2 / タイプ: protein_or_peptide / ID: 2 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 18.028131 KDa |
| 組換発現 | 生物種:  Trichoplusia ni (イラクサキンウワバ) Trichoplusia ni (イラクサキンウワバ) |
| 配列 | 文字列: MSGGLLKALR SDSYVELSQY RDQHFRGDNE EQEKLLKKSC TLYVGNLSFY TTEEQIYELF SKSGDIKKII MGLDKMKKTA CGFCFVEYY SRADAENAMR YINGTRLDDR IIRTDWDAGF KEGRQYGRGR SGGQVRDEYR QDYDAGRGGY GKLAQNQ UniProtKB: Nuclear cap-binding protein subunit 2 |
-分子 #3: RNA and export factor binding protein 2
| 分子 | 名称: RNA and export factor binding protein 2 / タイプ: protein_or_peptide / ID: 3 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  |
| 分子量 | 理論値: 17.542857 KDa |
| 組換発現 | 生物種:  |
| 配列 | 文字列: GAMGSMADKM DMSLDDIIKL NRNQRRVNRG GGPRRNRPAI ARGGRNRPAP YSRPKPLPDK WQHDLFDSGC GGGEGVETGA KLLVSNLDF GVSDADIQEL FAEFGTLKKA AVDYDRSGRS LGTADVHFER RADALKAMKQ YKGVPLDGRP MDIQLVTSQI D UniProtKB: RNA and export factor binding protein 2 |
-分子 #4: 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE
| 分子 | 名称: 7N-METHYL-8-HYDROGUANOSINE-5'-DIPHOSPHATE / タイプ: ligand / ID: 4 / コピー数: 1 / 式: M7G |
|---|---|
| 分子量 | 理論値: 458.235 Da |
| Chemical component information | 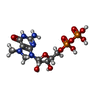 ChemComp-M7G: |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 8 |
|---|---|
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | TFS GLACIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: FEI FALCON IV (4k x 4k) 平均電子線量: 52.0 e/Å2 |
| 電子線 | 加速電圧: 200 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 2.0 µm / 最小 デフォーカス(公称値): 1.0 µm |
 ムービー
ムービー コントローラー
コントローラー
















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)