[English] 日本語
 Yorodumi
Yorodumi- EMDB-40228: Bacteriophage T4 capsid shell containing 9DE insertions into the ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Bacteriophage T4 capsid shell containing 9DE insertions into the gp23* major capsid protein subunits | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | Bacteriophage T4 / capsid shell / phage display / decorated capsid / phage head / VIRUS LIKE PARTICLE | ||||||||||||
| Function / homology | Capsid vertex protein / Major capsid protein, Myoviridae / Major capsid protein Gp23 / Capsid protein, T4-like bacteriophage-like / T=13 icosahedral viral capsid / viral capsid / Major capsid protein / Capsid vertex protein Function and homology information Function and homology information | ||||||||||||
| Biological species |  Tequatrovirus T4 / Tequatrovirus T4 /  Enterobacteria phage T4 (virus) Enterobacteria phage T4 (virus) | ||||||||||||
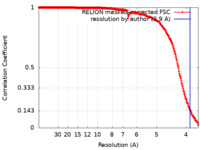
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | ||||||||||||
 Authors Authors | Fokine A / Rao VB | ||||||||||||
| Funding support |  United States, 3 items United States, 3 items
| ||||||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Design of bacteriophage T4-based artificial viral vectors for human genome remodeling. Authors: Jingen Zhu / Himanshu Batra / Neeti Ananthaswamy / Marthandan Mahalingam / Pan Tao / Xiaorong Wu / Wenzheng Guo / Andrei Fokine / Venigalla B Rao /  Abstract: Designing artificial viral vectors (AVVs) programmed with biomolecules that can enter human cells and carry out molecular repairs will have broad applications. Here, we describe an assembly-line ...Designing artificial viral vectors (AVVs) programmed with biomolecules that can enter human cells and carry out molecular repairs will have broad applications. Here, we describe an assembly-line approach to build AVVs by engineering the well-characterized structural components of bacteriophage T4. Starting with a 120 × 86 nm capsid shell that can accommodate 171-Kbp DNA and thousands of protein copies, various combinations of biomolecules, including DNAs, proteins, RNAs, and ribonucleoproteins, are externally and internally incorporated. The nanoparticles are then coated with cationic lipid to enable efficient entry into human cells. As proof of concept, we assemble a series of AVVs designed to deliver full-length dystrophin gene or perform various molecular operations to remodel human genome, including genome editing, gene recombination, gene replacement, gene expression, and gene silencing. These large capacity, customizable, multiplex, and all-in-one phage-based AVVs represent an additional category of nanomaterial that could potentially transform gene therapies and personalized medicine. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_40228.map.gz emd_40228.map.gz | 1.3 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-40228-v30.xml emd-40228-v30.xml emd-40228.xml emd-40228.xml | 18.3 KB 18.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_40228_fsc.xml emd_40228_fsc.xml | 26.6 KB | Display |  FSC data file FSC data file |
| Images |  emd_40228.png emd_40228.png | 253.5 KB | ||
| Masks |  emd_40228_msk_1.map emd_40228_msk_1.map | 1.4 GB |  Mask map Mask map | |
| Filedesc metadata |  emd-40228.cif.gz emd-40228.cif.gz | 6.4 KB | ||
| Others |  emd_40228_half_map_1.map.gz emd_40228_half_map_1.map.gz emd_40228_half_map_2.map.gz emd_40228_half_map_2.map.gz | 1.1 GB 1.1 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-40228 http://ftp.pdbj.org/pub/emdb/structures/EMD-40228 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40228 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-40228 | HTTPS FTP |
-Validation report
| Summary document |  emd_40228_validation.pdf.gz emd_40228_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_40228_full_validation.pdf.gz emd_40228_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_40228_validation.xml.gz emd_40228_validation.xml.gz | 31.9 KB | Display | |
| Data in CIF |  emd_40228_validation.cif.gz emd_40228_validation.cif.gz | 43.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40228 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40228 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40228 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-40228 | HTTPS FTP |
-Related structure data
| Related structure data |  8gmoMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_40228.map.gz / Format: CCP4 / Size: 1.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_40228.map.gz / Format: CCP4 / Size: 1.4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.85 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_40228_msk_1.map emd_40228_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_40228_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_40228_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Enterobacteria phage T4
| Entire | Name:  Enterobacteria phage T4 (virus) Enterobacteria phage T4 (virus) |
|---|---|
| Components |
|
-Supramolecule #1: Enterobacteria phage T4
| Supramolecule | Name: Enterobacteria phage T4 / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all / NCBI-ID: 10665 / Sci species name: Enterobacteria phage T4 / Virus type: VIRUS-LIKE PARTICLE / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes |
|---|---|
| Host (natural) | Organism:  |
-Macromolecule #1: Mature major capsid protein
| Macromolecule | Name: Mature major capsid protein / type: protein_or_peptide / ID: 1 / Number of copies: 93 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Tequatrovirus T4 Tequatrovirus T4 |
| Molecular weight | Theoretical: 49.790727 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AEIGGDHGYN ATNIAAGQTS GAVTQIGPAV MGMVRRAIPN LIAFDICGVQ PMNSPTGQVF ALRAVYGKDP VAAGAKEAFH PMYGPDAMF SGQGAAKKFP ALAASTQTTV GDIYTHFFQE TGTVYLQASV QVTIDAGDED EDEDEDATDA AKLDAEIKKQ M EAGALVEI ...String: AEIGGDHGYN ATNIAAGQTS GAVTQIGPAV MGMVRRAIPN LIAFDICGVQ PMNSPTGQVF ALRAVYGKDP VAAGAKEAFH PMYGPDAMF SGQGAAKKFP ALAASTQTTV GDIYTHFFQE TGTVYLQASV QVTIDAGDED EDEDEDATDA AKLDAEIKKQ M EAGALVEI AEGMATSIAE LQEGFNGSTD NPWNEMGFRI DKQVIEAKSR QLKAAYSIEL AQDLRAVHGM DADAELSGIL AT EIMLEIN REVVDWINYS AQVGKSGMTL TPGSKAGVFD FQDPIDIRGA RWAGESFKAL LFQIDKEAVE IARQTGRGEG NFI IASRNV VNVLASVDTG ISYAAQGLAT GFSTDTTKSV FAGVLGGKYR VYIDQYAKQD YFTVGYKGPN EMDAGIYYAP YVAL TPLRG SDPKNFQPVM GFKTRYGIGI NPFAESAAQA PASRIQSGMP SILNSLGKNA YFRRVYVKGI UniProtKB: Major capsid protein |
-Macromolecule #2: Mature capsid vertex protein
| Macromolecule | Name: Mature capsid vertex protein / type: protein_or_peptide / ID: 2 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Tequatrovirus T4 Tequatrovirus T4 |
| Molecular weight | Theoretical: 45.594375 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: STTTNSNSIG RPNLVALTRA TTKLIYSDIV ATQRTNQPVA AFYGIKYLNP DNEFTFKTGA TYAGEAGYVD REQITELTEE SKLTLNKGD LFKYNNIVYK VLEDTPFATI EESDLELALQ IAIVLLKVRL FSDAASTSKF ESSDSEIADA RFQINKWQTA V KSRKLKTG ...String: STTTNSNSIG RPNLVALTRA TTKLIYSDIV ATQRTNQPVA AFYGIKYLNP DNEFTFKTGA TYAGEAGYVD REQITELTEE SKLTLNKGD LFKYNNIVYK VLEDTPFATI EESDLELALQ IAIVLLKVRL FSDAASTSKF ESSDSEIADA RFQINKWQTA V KSRKLKTG ITVELAQDLE ANGFDAPNFL EDLLATEMAD EINKDILQSL ITVSKRYKVT GITDSGFIDL SYASAPEAGR SL YRMVCEM VSHIQKESTY TATFCVASAR AAAILAASGW LKHKPEDDKY LSQNAYGFLA NGLPLYCDTN SPLDYVIVGV VEN IGEKEI VGSIFYAPYT EGLDLDDPEH VGAFKVVVDP ESLQPSIGLL VRYALSANPY TVAKDEKEAR IIDGGDMDKM AGRS DLSVL LGVKLPKIII UniProtKB: Capsid vertex protein |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Model: PELCO Ultrathin Carbon with Lacey Carbon / Material: COPPER / Mesh: 400 / Support film - Material: CARBON / Support film - topology: LACEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 60 sec. |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 36.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.6 µm / Nominal magnification: 64000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)