[English] 日本語
 Yorodumi
Yorodumi- EMDB-38530: Cryo-EM structure of GPR30-Gq complex structure in the presence o... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of GPR30-Gq complex structure in the presence of fulvestrant | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / estrogen / GPR30 / Gq / MEMBRANE PROTEIN / MEMBRANE PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationnegative regulation of leukocyte activation / nuclear fragmentation involved in apoptotic nuclear change / positive regulation of cardiac vascular smooth muscle cell differentiation / negative regulation of cell cycle process / keratin filament / positive regulation of inositol trisphosphate biosynthetic process / positive regulation of uterine smooth muscle contraction / negative regulation of lipid biosynthetic process / apoptotic chromosome condensation / steroid hormone binding ...negative regulation of leukocyte activation / nuclear fragmentation involved in apoptotic nuclear change / positive regulation of cardiac vascular smooth muscle cell differentiation / negative regulation of cell cycle process / keratin filament / positive regulation of inositol trisphosphate biosynthetic process / positive regulation of uterine smooth muscle contraction / negative regulation of lipid biosynthetic process / apoptotic chromosome condensation / steroid hormone binding / cellular response to mineralocorticoid stimulus / positive regulation of G protein-coupled receptor signaling pathway / positive regulation of endothelial cell apoptotic process / positive regulation of extrinsic apoptotic signaling pathway / dendritic spine membrane / cellular response to peptide hormone stimulus / positive regulation of neurotransmitter secretion / positive regulation of neurogenesis / positive regulation of epidermal growth factor receptor signaling pathway / G protein-coupled estrogen receptor activity / nuclear estrogen receptor activity / positive regulation of release of cytochrome c from mitochondria / : / negative regulation of fat cell differentiation / dendritic spine head / presynaptic active zone / negative regulation of vascular associated smooth muscle cell proliferation / steroid hormone receptor signaling pathway / negative regulation of phosphatidylinositol 3-kinase/protein kinase B signal transduction / neuronal action potential / nuclear receptor-mediated steroid hormone signaling pathway / axon terminus / regulation of cytosolic calcium ion concentration / steroid binding / hippocampal mossy fiber to CA3 synapse / Peptide ligand-binding receptors / positive regulation of release of sequestered calcium ion into cytosol / dendritic shaft / cellular response to estradiol stimulus / G protein-coupled receptor activity / positive regulation of protein localization to plasma membrane / cellular response to glucose stimulus / mitochondrial membrane / trans-Golgi network / cytoplasmic vesicle membrane / positive regulation of insulin secretion / Olfactory Signaling Pathway / Activation of the phototransduction cascade / adenylate cyclase-activating G protein-coupled receptor signaling pathway / recycling endosome / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / negative regulation of ERK1 and ERK2 cascade / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / negative regulation of inflammatory response / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / vasodilation / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / sensory perception of taste / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / cellular response to prostaglandin E stimulus / Inactivation, recovery and regulation of the phototransduction cascade / G-protein beta-subunit binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / cellular response to tumor necrosis factor / GTPase binding / nuclear envelope / retina development in camera-type eye / presynaptic membrane / nervous system development / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / positive regulation of cytosolic calcium ion concentration / G alpha (i) signalling events / fibroblast proliferation Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / synthetic construct (others) Homo sapiens (human) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Liu H / Xu P / Xu HE | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2024 Journal: Cell Res / Year: 2024Title: Structural and functional evidence that GPR30 is not a direct estrogen receptor. Authors: Heng Liu / Shimeng Guo / Antao Dai / Peiyu Xu / Xin Li / Sijie Huang / Xinheng He / Kai Wu / Xinyue Zhang / Dehua Yang / Xin Xie / H Eric Xu /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38530.map.gz emd_38530.map.gz | 28.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38530-v30.xml emd-38530-v30.xml emd-38530.xml emd-38530.xml | 19.9 KB 19.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38530.png emd_38530.png | 40.8 KB | ||
| Masks |  emd_38530_msk_1.map emd_38530_msk_1.map | 30.5 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-38530.cif.gz emd-38530.cif.gz | 6.4 KB | ||
| Others |  emd_38530_half_map_1.map.gz emd_38530_half_map_1.map.gz emd_38530_half_map_2.map.gz emd_38530_half_map_2.map.gz | 23.3 MB 23.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38530 http://ftp.pdbj.org/pub/emdb/structures/EMD-38530 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38530 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38530 | HTTPS FTP |
-Validation report
| Summary document |  emd_38530_validation.pdf.gz emd_38530_validation.pdf.gz | 869.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_38530_full_validation.pdf.gz emd_38530_full_validation.pdf.gz | 868.9 KB | Display | |
| Data in XML |  emd_38530_validation.xml.gz emd_38530_validation.xml.gz | 10.5 KB | Display | |
| Data in CIF |  emd_38530_validation.cif.gz emd_38530_validation.cif.gz | 12.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38530 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38530 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38530 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-38530 | HTTPS FTP |
-Related structure data
| Related structure data |  8xoiMC  8xofC  8xogC  8xohC  8xojC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_38530.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38530.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.071 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_38530_msk_1.map emd_38530_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
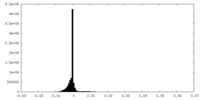
| Density Histograms |
-Half map: #2
| File | emd_38530_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
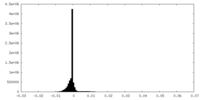
| Density Histograms |
-Half map: #1
| File | emd_38530_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
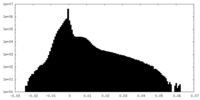
| Density Histograms |
- Sample components
Sample components
-Entire : GPR30-Gq complex
| Entire | Name: GPR30-Gq complex |
|---|---|
| Components |
|
-Supramolecule #1: GPR30-Gq complex
| Supramolecule | Name: GPR30-Gq complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: GPR30-Gq
| Supramolecule | Name: GPR30-Gq / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2, #4-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: ScFv16
| Supramolecule | Name: ScFv16 / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
-Macromolecule #1: Guanine nucleotide-binding protein G(q) subunit alpha-q
| Macromolecule | Name: Guanine nucleotide-binding protein G(q) subunit alpha-q type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.632219 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMIE KQLQKDKQVY RRTLRLLLLG ADNSGKSTIV KQMRIYHVNG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMIE KQLQKDKQVY RRTLRLLLLG ADNSGKSTIV KQMRIYHVNG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTSGIFETK FQVDKVNFHM FDVGAQRDER RKWIQCFNDV TAIIFVVDSS DTNRLQEALN DF DSIWNNR WLRTISVILF LNKQDLLAEK VLAGKSKIED YFPEFARYTT PEDATPEPGE DPRVTRAKYF IRKEFVDIST ASG DGRHIC YPHFTCAVDT ENARRIFNDC KDIILQMNLR EYNLV |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.744371 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI ...String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI VTSSGDTTCA LWDIETGQQT TTFTGHTGDV MSLSLAPDTR LFVSGACDAS AKLWDVREGM CRQTFTGHES DI NAICFFP NGNAFATGSD DATCRLFDLR ADQELMTYSH DNIICGITSV SFSKSGRLLL AGYDDFNCNV WDALKADRAG VLA GHDNRV SCLGVTDDGM AVATGSWDSF LKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 26.293299 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS AGGGGSGGGG SGGGGSSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYYA DTVKGRFTIS RDDPKNTLFL QMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS AGGGGSGGGG SGGGGSSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: G-protein coupled estrogen receptor 1
| Macromolecule | Name: G-protein coupled estrogen receptor 1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 42.291457 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDVTSQARGV GLEMYPGTAQ PAAPNTTSPE LNLSHPLLGT ALANGTGELS EHQQYVIGLF LSCLYTIFLF PIGFVGNILI LVVNISFRE KMTIPDLYFI NLAVADLILV ADSLIEVFNL HERYYDIAVL CTFMSLFLQV NMYSSVFFLT WMSFDRYIAL A RAMRCSLF ...String: MDVTSQARGV GLEMYPGTAQ PAAPNTTSPE LNLSHPLLGT ALANGTGELS EHQQYVIGLF LSCLYTIFLF PIGFVGNILI LVVNISFRE KMTIPDLYFI NLAVADLILV ADSLIEVFNL HERYYDIAVL CTFMSLFLQV NMYSSVFFLT WMSFDRYIAL A RAMRCSLF RTKHHARLSC GLIWMASVSA TLVPFTAVHL QHTDEACFCF ADVREVQWLE VTLGFIVPFA IIGLCYSLIV RV LVRAHRH RGLRPRRQKA LRMILAVVLV FFVCWLPENV FISVHLLQRT QPGAAPCKQS FRHAHPLTGH IVNLAAFSNS CLN PLIYSF LGETFRDKLR LYIEQKTNLP ALNRFCHAAL KAVIPDSTEQ SDVRFSSAV UniProtKB: G-protein coupled estrogen receptor 1 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DARK FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.2 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 536691 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller






























 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)