+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | ZIKV rsNS1 in complex with Fab AA12 and anti-fab nanobody | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | antibody / flavivirus / zika / cryoEM / non structural protein 1 / ns1 / VIRAL PROTEIN / VIRAL PROTEIN-IMMUNE SYSTEM complex | |||||||||
| Function / homology |  Function and homology information Function and homology informationflavivirin / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of host TYK2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / double-stranded RNA binding / nucleoside-triphosphate phosphatase / viral capsid / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / symbiont-mediated suppression of host toll-like receptor signaling pathway / mRNA (guanine-N7)-methyltransferase ...flavivirin / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of host TYK2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT2 activity / symbiont-mediated suppression of host JAK-STAT cascade via inhibition of STAT1 activity / double-stranded RNA binding / nucleoside-triphosphate phosphatase / viral capsid / symbiont-mediated suppression of host cytoplasmic pattern recognition receptor signaling pathway via inhibition of TBK1 activity / symbiont-mediated suppression of host toll-like receptor signaling pathway / mRNA (guanine-N7)-methyltransferase / methyltransferase cap1 / clathrin-dependent endocytosis of virus by host cell / mRNA (nucleoside-2'-O-)-methyltransferase activity / mRNA 5'-cap (guanine-N7-)-methyltransferase activity / RNA helicase activity / host cell perinuclear region of cytoplasm / protein dimerization activity / host cell endoplasmic reticulum membrane / RNA helicase / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / induction by virus of host autophagy / RNA-directed RNA polymerase / viral RNA genome replication / serine-type endopeptidase activity / RNA-dependent RNA polymerase activity / virus-mediated perturbation of host defense response / fusion of virus membrane with host endosome membrane / lipid binding / viral envelope / host cell nucleus / GTP binding / virion attachment to host cell / structural molecule activity / virion membrane / ATP hydrolysis activity / proteolysis / extracellular region / ATP binding / membrane / metal ion binding Similarity search - Function | |||||||||
| Biological species |   Zika virus / Zika virus /  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
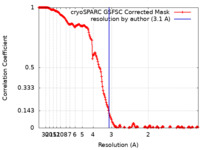
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Chew BLA / Luo D | |||||||||
| Funding support |  Singapore, 1 items Singapore, 1 items
| |||||||||
 Citation Citation |  Journal: Npj Viruses / Year: 2024 Journal: Npj Viruses / Year: 2024Title: Structural basis of Zika virus NS1 multimerization and human antibody recognition Authors: Chew BLA / Ngoh AQ / Phoo WW / Weng MJG / Sheng HJ / Chan KWK / Tan EYJ / Gelbart T / Xu C / Tan GS / Vasudevan SG / Luo D | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37670.map.gz emd_37670.map.gz | 567.3 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37670-v30.xml emd-37670-v30.xml emd-37670.xml emd-37670.xml | 17.1 KB 17.1 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37670_fsc.xml emd_37670_fsc.xml | 17.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_37670.png emd_37670.png | 142.7 KB | ||
| Filedesc metadata |  emd-37670.cif.gz emd-37670.cif.gz | 6 KB | ||
| Others |  emd_37670_half_map_1.map.gz emd_37670_half_map_1.map.gz emd_37670_half_map_2.map.gz emd_37670_half_map_2.map.gz | 557.9 MB 557.8 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37670 http://ftp.pdbj.org/pub/emdb/structures/EMD-37670 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37670 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37670 | HTTPS FTP |
-Validation report
| Summary document |  emd_37670_validation.pdf.gz emd_37670_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37670_full_validation.pdf.gz emd_37670_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_37670_validation.xml.gz emd_37670_validation.xml.gz | 27.1 KB | Display | |
| Data in CIF |  emd_37670_validation.cif.gz emd_37670_validation.cif.gz | 35.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37670 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37670 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37670 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37670 | HTTPS FTP |
-Related structure data
| Related structure data |  8wnpMC  8wn8C  8wnuC  8wo0C  8wo4C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37670.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37670.map.gz / Format: CCP4 / Size: 600.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.68 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_37670_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_37670_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ZIKV rsNS1 in complex with Fab AA12 and anti-fab nanobody
| Entire | Name: ZIKV rsNS1 in complex with Fab AA12 and anti-fab nanobody |
|---|---|
| Components |
|
-Supramolecule #1: ZIKV rsNS1 in complex with Fab AA12 and anti-fab nanobody
| Supramolecule | Name: ZIKV rsNS1 in complex with Fab AA12 and anti-fab nanobody type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:   Zika virus Zika virus |
-Macromolecule #1: VL Chains
| Macromolecule | Name: VL Chains / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.385945 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SDIQMTQSPL SLSASVGDRV TITCRTSQSI SSYLNWYQQK PGKAPKLLIY AASSLQSGVP SRFSGSGSGT DFTLTISSLQ PEDFATYYC QQTYSTPLTF GGGTKVEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD ...String: SDIQMTQSPL SLSASVGDRV TITCRTSQSI SSYLNWYQQK PGKAPKLLIY AASSLQSGVP SRFSGSGSGT DFTLTISSLQ PEDFATYYC QQTYSTPLTF GGGTKVEIKR TVAAPSVFIF PPSDEQLKSG TASVVCLLNN FYPREAKVQW KVDNALQSGN S QESVTEQD SKDSTYSLSS TLTLSKADYE KHKVYACEVT HQGLSSPVTK SFNRGEC |
-Macromolecule #2: VH Chains
| Macromolecule | Name: VH Chains / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 23.824621 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: SEVQLVESGG GLIQPGGSLR LSCAASGFTV SSNYMSWVRQ APGKGLEWVS VIYSGGSTYY ADSVKGRFTI SRDNSKNTLY LQMNSLRAE DTAVYYCARD RRGFDYWGQG TMVTVSSAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF PEPVTVSWNS G ALTSGVHT ...String: SEVQLVESGG GLIQPGGSLR LSCAASGFTV SSNYMSWVRQ APGKGLEWVS VIYSGGSTYY ADSVKGRFTI SRDNSKNTLY LQMNSLRAE DTAVYYCARD RRGFDYWGQG TMVTVSSAST KGPSVFPLAP SSKSTSGGTA ALGCLVKDYF PEPVTVSWNS G ALTSGVHT FPAVLQSSGL YSLSSVVTVP SSSLGTQTYI CNVNHKPSNT KVDKKVEPKS CDKTHT |
-Macromolecule #3: Non-structural protein 1
| Macromolecule | Name: Non-structural protein 1 / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:   Zika virus Zika virus |
| Molecular weight | Theoretical: 40.135375 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DVGCSVDFSK KETRCGTGVF IYNDVEAWRD RYKYHPDSPR RLAAAVKQAW EEGICGISSV SRMENIMWKS VEGELNAILE ENGVQLTVV VGSVKNPMWR GPQRLPVPVN ELPHGWKAWG KSYFVRAAKT NNSFVVDGDT LKECPLEHRA WNSFLVEDHG F GVFHTSVW ...String: DVGCSVDFSK KETRCGTGVF IYNDVEAWRD RYKYHPDSPR RLAAAVKQAW EEGICGISSV SRMENIMWKS VEGELNAILE ENGVQLTVV VGSVKNPMWR GPQRLPVPVN ELPHGWKAWG KSYFVRAAKT NNSFVVDGDT LKECPLEHRA WNSFLVEDHG F GVFHTSVW LKVREDYSLE CDPAVIGTAV KGREAAHSDL GYWIESEKND TWRLKRAHLI EMKTCEWPKS HTLWTDGVEE SD LIIPKSL AGPLSHHNTR EGYRTQVKGP WHSEELEIRF EECPGTKVYV EETCGTRGPS LRSTTASGRV IEEWCCRECT MPP LSFRAK DGCWYGMEIR PRKEPESNLV RSMVTA UniProtKB: Genome polyprotein |
-Macromolecule #4: Anti-fab nanobody
| Macromolecule | Name: Anti-fab nanobody / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.118386 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: VQLQESGGGL VQPGGSLRLS CAASGRTISR YAMSWFRQAP GKEREFVAVA RRSGDGAFYA DSVQGRFTVS RDDAKNTVYL QMNSLKPED TAVYYCAIDS DTFYSGSYDY WGQGTQVTVS S |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 2 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: OTHER / Imaging mode: OTHER / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Protocol: AB INITIO MODEL |
|---|---|
| Output model |  PDB-8wnp: |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)