[English] 日本語
 Yorodumi
Yorodumi- EMDB-37499: Cryo-EM structure of CRISPR-Csm effector complex from Mycobacteri... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of CRISPR-Csm effector complex from Mycobacterium canettii | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Csm effector complex / Mycobacterium canettii / Cryo-EM structure / Csm1 / Csm2 / Csm3 / Csm4 / Csm5 / RNA BINDING PROTEIN / RNA BINDING PROTEIN-RNA complex | |||||||||
| Function / homology | : / : / : / : / :  Function and homology information Function and homology information | |||||||||
| Biological species |  Mycobacterium canettii (bacteria) Mycobacterium canettii (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.73 Å | |||||||||
 Authors Authors | Huo Y / Ma X / Jiang T | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Int J Biol Macromol / Year: 2024 Journal: Int J Biol Macromol / Year: 2024Title: Type-III-A structure of mycobacteria CRISPR-Csm complexes involving atypical crRNAs. Authors: Hongtai Zhang / Mingmin Shi / Xiaoli Ma / Mengxi Liu / Nenhan Wang / Qiuhua Lu / Zekai Li / Yanfeng Zhao / Hongshen Zhao / Hong Chen / Huizhi Zhang / Tao Jiang / Songying Ouyang / Yangao Huo / Lijun Bi /  Abstract: Tuberculosis (TB), a leading cause of mortality globally, is a chronic infectious disease caused by Mycobacterium tuberculosis that primarily infiltrates the lung. The mature crRNAs in M. ...Tuberculosis (TB), a leading cause of mortality globally, is a chronic infectious disease caused by Mycobacterium tuberculosis that primarily infiltrates the lung. The mature crRNAs in M. tuberculosis transcribed from the Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) locus exhibit an atypical structure featured with 5' and 3' repeat tags at both ends of the intact crRNA, in contrast to typical Type-III-A crRNAs that possess 5' repeat tags and partial crRNA sequences. However, this structural peculiarity particularly concerning the specific binding characteristics of the 3' repeat end within the mature crRNA within the Csm complex, has not been comprehensively elucidated. Here, our Mycobacteria CRISPR-Csm complexes structure represents the largest Csm complex reported to date. It incorporates an atypical Type-III-A CRISPR RNA (crRNA) (46 nt) with 5' 8-nt and 3' 4-nt repeat sequences in the stoichiometry of Mycobacteria Csm12345 The PAM-independent single-stranded RNAs (ssRNAs) are the most suitable substrate for the Csm complex. The 3'-repeat end trimming of mature crRNA was not necessary for its cleavage activity in Type-III-A Csm complex. Our work broadens our understanding of the Type-III-A Csm complex and identifies another mature crRNA processing mechanism in the Type-III-A CRISPR-Cas system based on structural biology. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37499.map.gz emd_37499.map.gz | 202.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37499-v30.xml emd-37499-v30.xml emd-37499.xml emd-37499.xml | 20.1 KB 20.1 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37499.png emd_37499.png | 30.3 KB | ||
| Filedesc metadata |  emd-37499.cif.gz emd-37499.cif.gz | 6.8 KB | ||
| Others |  emd_37499_half_map_1.map.gz emd_37499_half_map_1.map.gz emd_37499_half_map_2.map.gz emd_37499_half_map_2.map.gz | 171.8 MB 171.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37499 http://ftp.pdbj.org/pub/emdb/structures/EMD-37499 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37499 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37499 | HTTPS FTP |
-Validation report
| Summary document |  emd_37499_validation.pdf.gz emd_37499_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37499_full_validation.pdf.gz emd_37499_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_37499_validation.xml.gz emd_37499_validation.xml.gz | 14.9 KB | Display | |
| Data in CIF |  emd_37499_validation.cif.gz emd_37499_validation.cif.gz | 17.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37499 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37499 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37499 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37499 | HTTPS FTP |
-Related structure data
| Related structure data |  8wfxMC  8x5dC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_37499.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37499.map.gz / Format: CCP4 / Size: 216 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||||||||||||||||||
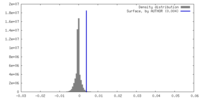
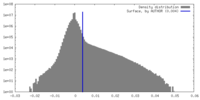
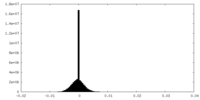
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_37499_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
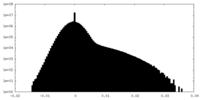
| Density Histograms |
-Half map: #1
| File | emd_37499_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Csm effector complex
| Entire | Name: Csm effector complex |
|---|---|
| Components |
|
-Supramolecule #1: Csm effector complex
| Supramolecule | Name: Csm effector complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  Mycobacterium canettii (bacteria) Mycobacterium canettii (bacteria) |
-Macromolecule #1: CRISPR system Cms protein Csm4
| Macromolecule | Name: CRISPR system Cms protein Csm4 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium canettii (bacteria) Mycobacterium canettii (bacteria) |
| Molecular weight | Theoretical: 32.26477 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MNSRLFRFDV VRTHFGDHGL ESSTIGCPAD TLYSALCVEA LRMGGQQLLD ELVACSTLRL TDLLPYVGPD YLVPKPLKSV RSDSSSLQK KLAKKIGFLP AAQLGSFLDG TADLDDLAAR QTKIGVHAVS AKAAIHNGKK DADPYRVGYF RFEPDAGLWL L ATGSESEL ...String: MNSRLFRFDV VRTHFGDHGL ESSTIGCPAD TLYSALCVEA LRMGGQQLLD ELVACSTLRL TDLLPYVGPD YLVPKPLKSV RSDSSSLQK KLAKKIGFLP AAQLGSFLDG TADLDDLAAR QTKIGVHAVS AKAAIHNGKK DADPYRVGYF RFEPDAGLWL L ATGSESEL GLLTRLLKGI SALGGERTSG FGAFIPTESE VPAALTPTTA AASLMTLTTS LPTDDELEQA LAGATYRLVK RS GFVASAT YAETPRRKRD IYKLAAGSVF SRPFQGGILD VSLGGNHPVY SYARPLFLAL PESAA UniProtKB: UNIPROTKB: G0TFC1 |
-Macromolecule #3: CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1...
| Macromolecule | Name: CRISPR system single-strand-specific deoxyribonuclease Cas10/Csm1 (subtype III-A) type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium canettii (bacteria) Mycobacterium canettii (bacteria) |
| Molecular weight | Theoretical: 91.957586 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MIPALIETVI GCLLHDIGKP VQRAALGYRG RHSAIGRAFV KKIWLRDGRN PSEFADEVYE PDIEVFDRRI LDAISYHHSS ALRTAAENG RLAADAPAYV AYIADNIAAG TDRRKADSDD GQGASTWDSD TPLYSVFNRF GADTANLTFA PEMLDDREPM N IPSARRIE ...String: MIPALIETVI GCLLHDIGKP VQRAALGYRG RHSAIGRAFV KKIWLRDGRN PSEFADEVYE PDIEVFDRRI LDAISYHHSS ALRTAAENG RLAADAPAYV AYIADNIAAG TDRRKADSDD GQGASTWDSD TPLYSVFNRF GADTANLTFA PEMLDDREPM N IPSARRIE FDKYRYTEIV NKLEAVLVDL ECSDTYLASL LNVLEATLSF VPSSTDASEV VDVSLFDHLK LTGALGACIW HY LQATGQS DFKSALFDKQ DTFYNEKAFL LTTFDVSGIQ DFIYTIHSSG AAKMLRARSF YLEMLTEHLI DELLARVGLS RAN LNYSGG GHAYLLLPNT EFARNSLEEF EREANEWLLE NFATWLFIAT GSVPLAANDL MRRPNESGPQ AHDRALRYSG LYRE LSEQL SAKKLARYSA DQLRELNSSD HDGQKGDREC SVCHTVNRTI KTINDLLLCS LCQALTAASQ QIQSESRRFL LISEE SSDG LPLPFGATLT FCSESGAKQA LQQPQTRRLY AKNKFFAGES LGTGLWVGDY VAQMEFGDYV KRASGIARLG VLRLDV DNL GQAFTHGFMK QGNGKFNTIS RTAAFSRMLS LFFRQHINYV LKHPKLRPLT GDDPERSRRA TIIYSGGDDV FVVGAWD DV IEFGIELRER FHEFTQGKLT VSAGIGMFPD KYPISVMARE VGDLEGAAKS LPGKNGVALF DPEFTFRWDE LLSKVIEE K YRHIADYFRC NEERGMAFIY KLLELLDQRG DRITEARWVY FLMRMRGPTD DTTRFQQFAN RLHQWFQDPS DAKQLKTAL HLYIYRNRKE ESE UniProtKB: UNIPROTKB: G0TFC4 |
-Macromolecule #4: CRISPR system Cms protein Csm5
| Macromolecule | Name: CRISPR system Cms protein Csm5 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium canettii (bacteria) Mycobacterium canettii (bacteria) |
| Molecular weight | Theoretical: 42.167949 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSQYLKPFEL TLRCLGPVFI GSGEKRTPKE YVASTSMVYF PDMERLYADV AAQGKSESFE EFMMNTGKAQ PDERFNEWIA ENGVKVSPK NHGGYGVKIG SIVPGRAHRG RDGQMIQEQR QLNDIHSFIK DVLGNPYVPG SSVKGMLRSI YLQSLVHQRT A QPVRVPGH ...String: MSQYLKPFEL TLRCLGPVFI GSGEKRTPKE YVASTSMVYF PDMERLYADV AAQGKSESFE EFMMNTGKAQ PDERFNEWIA ENGVKVSPK NHGGYGVKIG SIVPGRAHRG RDGQMIQEQR QLNDIHSFIK DVLGNPYVPG SSVKGMLRSI YLQSLVHQRT A QPVRVPGH QTREHRQYGE RFERKELRKS GRPNTRPQDA VNDLFQAIRV TDSPGLRTSD LLICQKMDVN VHGKPDGLPL FR ECLAPGT SISLRVVVDT SPTARGGWPA GERFLETLSD TVAFVNKARY AEYAAKYWDD DPQFGPIVYL GGGAGYRSKT FVT QQDDMA KVLDAQFPKI KHVAKTRDLG VSPLVLKLTK IGDKYYEMGQ CELSIRRAE UniProtKB: UNIPROTKB: G0TFC0 |
-Macromolecule #5: CRISPR system Cms protein Csm2
| Macromolecule | Name: CRISPR system Cms protein Csm2 / type: protein_or_peptide / ID: 5 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium canettii (bacteria) Mycobacterium canettii (bacteria) |
| Molecular weight | Theoretical: 15.006156 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSVIQDDYVK QAEQVIRGLP KKNGDFELTT TQLRVLLSLT AQLFDEAQLS SDQNLSPALR DKVQYLRVRF VYQAGREKAV RVFVERAGL LDELAQIGDS RDRLLKFCHY MEALVAYKKF LDPKETSKET E UniProtKB: UNIPROTKB: G0TFC3 |
-Macromolecule #6: CRISPR system Cms endoribonuclease Csm3
| Macromolecule | Name: CRISPR system Cms endoribonuclease Csm3 / type: protein_or_peptide / ID: 6 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium canettii (bacteria) Mycobacterium canettii (bacteria) |
| Molecular weight | Theoretical: 25.952537 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MITGYAKIEI TGTITVVTGL HIGAGDGFSA IGAVDKPVVR DPLTKWPMIP GTSLKGKVRT LLSRQYGADT ETFYRKPNDD PAQIRRLFG DTKEYKTGRL VFRDTKLTNK DDLEARGAKT LTEVKFENAI NRVTAKANPR QMERVIPGSE FAFSLVYEVS F GTPDEEQK ...String: MITGYAKIEI TGTITVVTGL HIGAGDGFSA IGAVDKPVVR DPLTKWPMIP GTSLKGKVRT LLSRQYGADT ETFYRKPNDD PAQIRRLFG DTKEYKTGRL VFRDTKLTNK DDLEARGAKT LTEVKFENAI NRVTAKANPR QMERVIPGSE FAFSLVYEVS F GTPDEEQK ASLPSSDEII EDFNAIARGL KLLELDYLGG SGTRGYGQVK FSDLKARAAV GTLDRSLLEM LNHELAAV UniProtKB: UNIPROTKB: G0TFC2 |
-Macromolecule #2: RNA (50-MER)
| Macromolecule | Name: RNA (50-MER) / type: rna / ID: 2 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  Mycobacterium canettii (bacteria) Mycobacterium canettii (bacteria) |
| Molecular weight | Theoretical: 16.001554 KDa |
| Sequence | String: ACGGAAACUU AAAACCGUGU UGCACUGCAA CCCGGAAUUC UUGCACGUCG |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.73 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 110023 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller


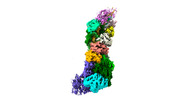

 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)