[English] 日本語
 Yorodumi
Yorodumi- EMDB-38066: The cryo-EM structure of the Mycobacterium tuberculosis CRISPR-Cs... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The cryo-EM structure of the Mycobacterium tuberculosis CRISPR-Csm complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Mycobacteria CRISPR-Csm complexes / RNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationendonuclease activity / defense response to virus / hydrolase activity / RNA binding Similarity search - Function | |||||||||
| Biological species |   Mycobacterium canettii (bacteria) Mycobacterium canettii (bacteria) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Liu MX / Li ZK | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation | Journal: FASEB J / Year: 2019 Title: Mycobacterium tuberculosis type III-A CRISPR/Cas system crRNA and its maturation have atypical features. Authors: Wenjing Wei / Shuai Zhang / Joy Fleming / Ying Chen / Zihui Li / Shanghua Fan / Yi Liu / Wei Wang / Ting Wang / Ying Liu / Baiguang Ren / Ming Wang / Jianjian Jiao / Yuanyuan Chen / Ying ...Authors: Wenjing Wei / Shuai Zhang / Joy Fleming / Ying Chen / Zihui Li / Shanghua Fan / Yi Liu / Wei Wang / Ting Wang / Ying Liu / Baiguang Ren / Ming Wang / Jianjian Jiao / Yuanyuan Chen / Ying Zhou / Yafeng Zhou / Shoujin Gu / Xiaoli Zhang / Li Wan / Tao Chen / Lin Zhou / Yong Chen / Xian-En Zhang / Chuanyou Li / Hongtai Zhang / Lijun Bi /  Abstract: Clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein (Cas) systems are prokaryotic adaptive immune systems against invading nucleic acids. CRISPR locus ...Clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein (Cas) systems are prokaryotic adaptive immune systems against invading nucleic acids. CRISPR locus variability has been exploited in evolutionary and epidemiological studies of Mycobacterium tuberculosis, the causative agent of tuberculosis, for over 20 yr, yet the biological function of this type III-A system is largely unexplored. Here, using cell biology and biochemical, mutagenic, and RNA-seq approaches, we show it is active in invader defense and has features atypical of type III-A systems: mature CRISPR RNA (crRNA) in its crRNA-CRISPR/Cas protein complex are of uniform length (∼71 nt) and appear not to be subject to 3'-end processing after Cas6 cleavage of repeat RNA 8 nt from its 3' end. crRNAs generated resemble mature crRNA in type I systems, having both 5' (8 nt) and 3' (28 nt) repeat tags. Cas6 cleavage of repeat RNA is ion dependent, and accurate cleavage depends on the presence of a 3' hairpin in the repeat RNA and the sequence of its stem base nucleotides. This study unveils further diversity among CRISPR/Cas systems and provides insight into the crRNA recognition mechanism in M. tuberculosis, providing a foundation for investigating the potential of a type III-A-based genome editing system.-Wei, W., Zhang, S., Fleming, J., Chen, Y., Li, Z., Fan, S., Liu, Y., Wang, W., Wang, T., Liu, Y., Ren, B., Wang, M., Jiao, J., Chen, Y., Zhou, Y., Zhou, Y., Gu, S., Zhang, X., Wan, L., Chen, T., Zhou, L., Chen, Y., Zhang, X.-E., Li, C., Zhang, H., Bi, L. Mycobacterium tuberculosis type III-A CRISPR/Cas system crRNA and its maturation have atypical features. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_38066.map.gz emd_38066.map.gz | 450.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-38066-v30.xml emd-38066-v30.xml emd-38066.xml emd-38066.xml | 22.3 KB 22.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_38066.png emd_38066.png | 14.9 KB | ||
| Filedesc metadata |  emd-38066.cif.gz emd-38066.cif.gz | 6.8 KB | ||
| Others |  emd_38066_half_map_1.map.gz emd_38066_half_map_1.map.gz emd_38066_half_map_2.map.gz emd_38066_half_map_2.map.gz | 442.4 MB 442.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-38066 http://ftp.pdbj.org/pub/emdb/structures/EMD-38066 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38066 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-38066 | HTTPS FTP |
-Related structure data
| Related structure data |  8x5dMC  8wfxC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_38066.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_38066.map.gz / Format: CCP4 / Size: 476.8 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.855 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_38066_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
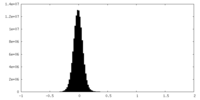
| Density Histograms |
-Half map: #2
| File | emd_38066_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
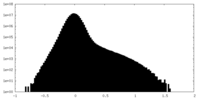
| Density Histograms |
- Sample components
Sample components
-Entire : Mycobacteria CRISPR-Csm complexes
| Entire | Name: Mycobacteria CRISPR-Csm complexes |
|---|---|
| Components |
|
-Supramolecule #1: Mycobacteria CRISPR-Csm complexes
| Supramolecule | Name: Mycobacteria CRISPR-Csm complexes / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: Csm2
| Supramolecule | Name: Csm2 / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #4 |
|---|---|
| Source (natural) | Organism:  Mycobacterium canettii (bacteria) Mycobacterium canettii (bacteria) |
-Macromolecule #1: CRISPR system Cms endoribonuclease Csm3
| Macromolecule | Name: CRISPR system Cms endoribonuclease Csm3 / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.147736 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAHMTTSYAK IEITGTLTVL TGLQIGAGDG FSAIGAVDKP VVRDPLSRLP MIPGTSLKGK VRTLLSRQYG ADTETFYRKP NEDHAHIRR LFGDTEEYMT GRLVFRDTKL TNKDDLEARG AKTLTEVKFE NAINRVTAKA NLRQMERVIP GSEFAFSLVY E VSFGTPGE ...String: MAHMTTSYAK IEITGTLTVL TGLQIGAGDG FSAIGAVDKP VVRDPLSRLP MIPGTSLKGK VRTLLSRQYG ADTETFYRKP NEDHAHIRR LFGDTEEYMT GRLVFRDTKL TNKDDLEARG AKTLTEVKFE NAINRVTAKA NLRQMERVIP GSEFAFSLVY E VSFGTPGE EQKASLPSSD EIIEDFNAIA RGLKLLELDY LGGSGTRGYG QVKFSNLKAR AAVGALDGSL LEKLNHELAA V UniProtKB: CRISPR system Cms endoribonuclease Csm3 |
-Macromolecule #2: CRISPR system Cms protein Csm5
| Macromolecule | Name: CRISPR system Cms protein Csm5 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 42.752129 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAHMNTYLKP FELTLRCLGP VFIGSGEKRT SKEYHVEGDR VYFPDMELLY ADIPAHKRKS FEAFVMNTDG AQATAPLKEW VEPNAVKLD PAKHRGYEVK IGSIEPRRAS RGRGGRMTRK KLTLNEIHAF IKDPLGRPYV PGSTVKGMLR SIYLQSLVHK R TAQPVRVP ...String: MAHMNTYLKP FELTLRCLGP VFIGSGEKRT SKEYHVEGDR VYFPDMELLY ADIPAHKRKS FEAFVMNTDG AQATAPLKEW VEPNAVKLD PAKHRGYEVK IGSIEPRRAS RGRGGRMTRK KLTLNEIHAF IKDPLGRPYV PGSTVKGMLR SIYLQSLVHK R TAQPVRVP GHQTREHRQY GERFERKELR KSGRPNTRPQ DAVNDLFQAI RVTDSPALRT SDLLICQKMD MNVHGKPDGL PL FRECLAP GTSISHRVVV DTSPTARGGW REGERFLETL AETAASVNQA RYAEYRAMYP GVNAIVGPIV YLGGGAGYRS KTF VTDQDD MAKVLDAQFG KVVKHVDKTR ELRVSPLVLK RTKIDNICYE MGQCELSIRR AE UniProtKB: CRISPR system Cms protein Csm5 |
-Macromolecule #4: Csm2
| Macromolecule | Name: Csm2 / type: protein_or_peptide / ID: 4 / Number of copies: 5 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycobacterium canettii (bacteria) Mycobacterium canettii (bacteria) |
| Molecular weight | Theoretical: 15.346579 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MAHMSVIQDD YVKQAEQVIR GLPKKNGDFE LTTTQLRVLL SLTAQLFDEA QLSSDQNLSP ALRDKVQYLR VRFVYQAGRE KAVRVFVER AGLLDELAQI GDSRDRLLKF CHYMEALVAY KKFLDPKETS KETE |
-Macromolecule #3: RNA (47-MER)
| Macromolecule | Name: RNA (47-MER) / type: rna / ID: 3 / Number of copies: 1 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 59.360578 KDa |
| Sequence | String: GUCGUCAGAC CCAAAACCCC GAGAGGGGAC GGAAACUUAA AACCGUGUUG CACUGCAACC CGGAAUUCUU GCACGUCGUC AGACCCAAA ACCCCGAGAG GGGACGGAAA CUUAAAACCG UGUUGCACUG CAACCCGGAA UUCUUGCACG UCGUCAGACC C AAAACCCC GAGAGGGGAC GGAAAC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.5 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)