[English] 日本語
 Yorodumi
Yorodumi- EMDB-37456: Structural organization of the palisade layer in isolated vaccini... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structural organization of the palisade layer in isolated vaccinia virus cores | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | poxvirus / assembly / core / palisade layer / A10 / VIRAL PROTEIN | |||||||||
| Function / homology | Poxvirus P4A / Poxvirus P4A protein / virion component / structural molecule activity / Major core protein OPG136 precursor Function and homology information Function and homology information | |||||||||
| Biological species |  Vaccinia virus (strain Western Reserve) / Vaccinia virus (strain Western Reserve) /  Vaccinia virus Western Reserve Vaccinia virus Western Reserve | |||||||||
| Method | subtomogram averaging / cryo EM / Resolution: 10.0 Å | |||||||||
 Authors Authors | Liu Y / Qu X / Duan M / Shi X / Liu S / Shi Y / Gao GF | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: Cryo-ET reveals A10 protein as a major component of the poxvirus palisade layer Authors: Liu Y / Qu X / Duan M / Shi X / Liu S / Shi Y / Gao GF | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37456.map.gz emd_37456.map.gz | 1016 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37456-v30.xml emd-37456-v30.xml emd-37456.xml emd-37456.xml | 17.3 KB 17.3 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_37456_fsc.xml emd_37456_fsc.xml | 4.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_37456.png emd_37456.png | 151.8 KB | ||
| Masks |  emd_37456_msk_1.map emd_37456_msk_1.map | 6.6 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-37456.cif.gz emd-37456.cif.gz | 6.4 KB | ||
| Others |  emd_37456_half_map_1.map.gz emd_37456_half_map_1.map.gz emd_37456_half_map_2.map.gz emd_37456_half_map_2.map.gz | 3.2 MB 3.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37456 http://ftp.pdbj.org/pub/emdb/structures/EMD-37456 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37456 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37456 | HTTPS FTP |
-Validation report
| Summary document |  emd_37456_validation.pdf.gz emd_37456_validation.pdf.gz | 691.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37456_full_validation.pdf.gz emd_37456_full_validation.pdf.gz | 691 KB | Display | |
| Data in XML |  emd_37456_validation.xml.gz emd_37456_validation.xml.gz | 8.2 KB | Display | |
| Data in CIF |  emd_37456_validation.cif.gz emd_37456_validation.cif.gz | 11.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37456 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37456 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37456 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37456 | HTTPS FTP |
-Related structure data
| Related structure data |  8wd7MC  8wdcC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_37456.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37456.map.gz / Format: CCP4 / Size: 6.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 4 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_37456_msk_1.map emd_37456_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
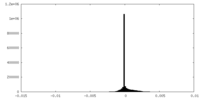
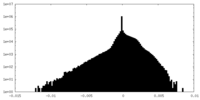
| Density Histograms |
-Half map: #1
| File | emd_37456_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_37456_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Vaccinia virus Western Reserve
| Entire | Name:  Vaccinia virus Western Reserve Vaccinia virus Western Reserve |
|---|---|
| Components |
|
-Supramolecule #1: Vaccinia virus Western Reserve
| Supramolecule | Name: Vaccinia virus Western Reserve / type: virus / ID: 1 / Parent: 0 / Macromolecule list: all Details: Vaccinia virus was grown in HeLa cells and purified. The resultant mature virions were treated with DTT and NP40 and then purified to yield viral cores. NCBI-ID: 696871 / Sci species name: Vaccinia virus Western Reserve / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: Yes / Virus empty: No |
|---|---|
| Host (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Core protein OPG136
| Macromolecule | Name: Core protein OPG136 / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vaccinia virus (strain Western Reserve) Vaccinia virus (strain Western Reserve) |
| Molecular weight | Theoretical: 71.073477 KDa |
| Sequence | String: MMPIKSIVTL DQLEDSEYLF RIVSTVLPHL CLDYKVCDQL KTTFVHPFDI LLNNSLGSVT KQDELQAAIS KLGINYLIDT TSRELKLFN VTLNAGNIDI INTPINISSE TNPIINTHSF YDLPPFTQHL LNIRLTDTEY RARFIGGYIK PDGSDSMDVL A EKKYPDLN ...String: MMPIKSIVTL DQLEDSEYLF RIVSTVLPHL CLDYKVCDQL KTTFVHPFDI LLNNSLGSVT KQDELQAAIS KLGINYLIDT TSRELKLFN VTLNAGNIDI INTPINISSE TNPIINTHSF YDLPPFTQHL LNIRLTDTEY RARFIGGYIK PDGSDSMDVL A EKKYPDLN FDNTYLFNIL YKDVINAPIK EFKAKIVNGV LSRQDFDNLI GVRQYITIQD RPRFDDAYNI ADAARHYGVN LN TLPLPNV DLTTMPTYKH LIMFEQYFIY TYDRVDIYYN GNKMLFDDEI INFTISMRYQ SLIPRLVDFF PDIPVNNNIV LHT RDPQNA AVNVTVALPN VQFVDINRNN KFFINFFNLL AKEQRSTAIK VTKSMFWDGM DYEEYKSKNL QDMMFINSTC YVFG LYNHN NTTYCSILSD IISAEKTPIR VCLLPRVVGG KTVTNLISET LKSISSMTIR EFPRKDKSIM HIGLSETGFM RFFQL LRLM ADKPHETAIK EVVMAYVGIK LGDKGSPYYI RKESYQDFIY LLFASMGFKV TTRRSIMGSN NISIISIRPR VTKQYI VAT LMKTSCSKNE AEKLITSAFD LLNFMVSVSD FRDYQSYRQY RNYCPRYFYA G UniProtKB: Major core protein OPG136 precursor |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 9 / Details: 1mM Tris pH 9 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: GOLD / Support film - Material: CARBON / Support film - topology: HOLEY |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
| Details | Vaccinia virus was grown in HeLa cells and purified. The resultant mature virions were treated with DTT and NP40 and then purified to yield viral cores. Isolated cores were used as a specimen. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Spherical aberration corrector: Microscope was equipped with a Cs corrector Chromatic aberration corrector: Microscope was equipped with a Cc corrector Energy filter - Name: TFS Selectris X / Energy filter - Slit width: 40 eV |
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 2.9 e/Å2 / Details: Dose rate 5.2 e-/pixel/s, EER (241 frames/sec) |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.0 mm / Nominal defocus max: 4.5 µm / Nominal defocus min: 1.6 µm / Nominal magnification: 53000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Initial model | Chain - Source name: AlphaFold / Chain - Initial model type: in silico model |
|---|---|
| Output model |  PDB-8wd7: |
 Movie
Movie Controller
Controller




 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)