+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the CHA-bound mTAAR1-Gq complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | CHA / mTAAR1 / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationG protein-coupled amine receptor activity / Amine ligand-binding receptors / G alpha (s) signalling events / trace-amine receptor activity / positive regulation of cAMP-mediated signaling / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor ...G protein-coupled amine receptor activity / Amine ligand-binding receptors / G alpha (s) signalling events / trace-amine receptor activity / positive regulation of cAMP-mediated signaling / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / sensory perception of taste / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / cellular response to prostaglandin E stimulus / Inactivation, recovery and regulation of the phototransduction cascade / G-protein beta-subunit binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / GTPase binding / retina development in camera-type eye / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / G alpha (q) signalling events / Ras protein signal transduction / cell population proliferation / Extra-nuclear estrogen signaling / G protein-coupled receptor signaling pathway / lysosomal membrane / GTPase activity / synapse / protein-containing complex binding / signal transduction / extracellular exosome / membrane / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.1 Å | |||||||||
 Authors Authors | Rong NK / Guo LL / Zhang MH / Li Q / Yang F / Sun JP | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Structural and signaling mechanisms of TAAR1 enabled preferential agonist design. Authors: Pan Shang / Naikang Rong / Jing-Jing Jiang / Jie Cheng / Ming-Hui Zhang / Dongwei Kang / Lei Qi / Lulu Guo / Gong-Ming Yang / Qun Liu / Zhenzhen Zhou / Xiao-Bing Li / Kong-Kai Zhu / Qing- ...Authors: Pan Shang / Naikang Rong / Jing-Jing Jiang / Jie Cheng / Ming-Hui Zhang / Dongwei Kang / Lei Qi / Lulu Guo / Gong-Ming Yang / Qun Liu / Zhenzhen Zhou / Xiao-Bing Li / Kong-Kai Zhu / Qing-Biao Meng / Xiang Han / Wenqi Yan / Yalei Kong / Lejin Yang / Xiaohui Wang / Dapeng Lei / Xin Feng / Xinyong Liu / Xiao Yu / Yue Wang / Qian Li / Zhen-Hua Shao / Fan Yang / Jin-Peng Sun /  Abstract: Trace amine-associated receptor 1 (TAAR1) senses a spectrum of endogenous amine-containing metabolites (EAMs) to mediate diverse psychological functions and is useful for schizophrenia treatment ...Trace amine-associated receptor 1 (TAAR1) senses a spectrum of endogenous amine-containing metabolites (EAMs) to mediate diverse psychological functions and is useful for schizophrenia treatment without the side effects of catalepsy. Here, we systematically profiled the signaling properties of TAAR1 activation and present nine structures of TAAR1-Gs/Gq in complex with EAMs, clinical drugs, and synthetic compounds. These structures not only revealed the primary amine recognition pocket (PARP) harboring the conserved acidic D for conserved amine recognition and "twin" toggle switch for receptor activation but also elucidated that targeting specific residues in the second binding pocket (SBP) allowed modulation of signaling preference. In addition to traditional drug-induced Gs signaling, Gq activation by EAM or synthetic compounds is beneficial to schizophrenia treatment. Our results provided a structural and signaling framework for molecular recognition by TAAR1, which afforded structural templates and signal clues for TAAR1-targeted candidate compounds design. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_37437.map.gz emd_37437.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-37437-v30.xml emd-37437-v30.xml emd-37437.xml emd-37437.xml | 19.6 KB 19.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_37437.png emd_37437.png | 112.4 KB | ||
| Filedesc metadata |  emd-37437.cif.gz emd-37437.cif.gz | 6.5 KB | ||
| Others |  emd_37437_half_map_1.map.gz emd_37437_half_map_1.map.gz emd_37437_half_map_2.map.gz emd_37437_half_map_2.map.gz | 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-37437 http://ftp.pdbj.org/pub/emdb/structures/EMD-37437 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37437 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-37437 | HTTPS FTP |
-Validation report
| Summary document |  emd_37437_validation.pdf.gz emd_37437_validation.pdf.gz | 758.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_37437_full_validation.pdf.gz emd_37437_full_validation.pdf.gz | 758.2 KB | Display | |
| Data in XML |  emd_37437_validation.xml.gz emd_37437_validation.xml.gz | 12.4 KB | Display | |
| Data in CIF |  emd_37437_validation.cif.gz emd_37437_validation.cif.gz | 14.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37437 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37437 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37437 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-37437 | HTTPS FTP |
-Related structure data
| Related structure data |  8wcbMC  8wc3C  8wc4C  8wc5C  8wc6C  8wc7C  8wc8C  8wc9C  8wcaC  8wccC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_37437.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_37437.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.92 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_37437_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_37437_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of the CHA-bound mTAAR1-Gq complex
| Entire | Name: Cryo-EM structure of the CHA-bound mTAAR1-Gq complex |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of the CHA-bound mTAAR1-Gq complex
| Supramolecule | Name: Cryo-EM structure of the CHA-bound mTAAR1-Gq complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Engineered G-alpha-q subunit
| Macromolecule | Name: Engineered G-alpha-q subunit / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.708383 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMIE KQLQKDKQVY RRTLRLLLLG ADNSGKSTIV KQMRIYHVNG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMIE KQLQKDKQVY RRTLRLLLLG ADNSGKSTIV KQMRIYHVNG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTSGIFETK FQVDKVNFHM FDVGAQRDER RKWIQCFNDV TAIIFVVDSS DYNRLQEALN DF KSIWNNR WLRTISVILF LNKQDLLAEK VLAGKSKIED YFPEFARYTT PEDATPEPGE DPRVTRAKYF IRKEFVDIST ASG DGRHIC YPHFTCAVDT ENARRIFNDC KDIILQMNLR EYNLV |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 37.915496 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD ...String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD TTCALWDIET GQQTTTFTGH TGDVMSLSLA PDTRLFVSGA CDASAKLWDV REGMCRQTFT GHESDINAIC FF PNGNAFA TGSDDATCRL FDLRADQELM TYSHDNIICG ITSVSFSKSG RLLLAGYDDF NCNVWDALKA DRAGVLAGHD NRV SCLGVT DDGMAVATGS WDSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: Trace amine-associated receptor 1
| Macromolecule | Name: Trace amine-associated receptor 1 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.656586 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHLCHAITNI SHRNSDWSRE VQASLYSLMS LIILATLVGN LIVIISISHF KQLHTPTNWL LHSMAIVDFL LGCLIMPCSM VRTVERCWY FGEILCKVHT STDIMLSSAS IFHLAFISID RYCAVCDPLR YKAKINISTI LVMILVSWSL PAVYAFGMIF L ELNLKGVE ...String: MHLCHAITNI SHRNSDWSRE VQASLYSLMS LIILATLVGN LIVIISISHF KQLHTPTNWL LHSMAIVDFL LGCLIMPCSM VRTVERCWY FGEILCKVHT STDIMLSSAS IFHLAFISID RYCAVCDPLR YKAKINISTI LVMILVSWSL PAVYAFGMIF L ELNLKGVE ELYRSQVSDL GGCSPFFSKV SGVLAFMTSF YIPGSVMLFV YYRIYFIAKG QARSINRTNV QVGLEGKSQA PQ SKETKAA KTLGIMVGVF LVCWCPFFLC TVLDPFLGYV IPPSLNDALY WFGYLNSALN PMVYAFFYPW FRRALKMVLL GKI FQKDSS RSKLFL UniProtKB: Trace amine-associated receptor 1 |
-Macromolecule #4: scFv16
| Macromolecule | Name: scFv16 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 30.363043 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFAVQ LVESGGGLVQ PGGSRKLSCS ASGFAFSSFG MHWVRQAPEK GLEWVAYIS SGSGTIYYAD TVKGRFTISR DDPKNTLFLQ MTSLRSEDTA MYYCVRSIYY YGSSPFDFWG QGTTLTVSAG G GGSGGGGS ...String: MLLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFAVQ LVESGGGLVQ PGGSRKLSCS ASGFAFSSFG MHWVRQAPEK GLEWVAYIS SGSGTIYYAD TVKGRFTISR DDPKNTLFLQ MTSLRSEDTA MYYCVRSIYY YGSSPFDFWG QGTTLTVSAG G GGSGGGGS GGGGSADIVM TQATSSVPVT PGESVSISCR SSKSLLHSNG NTYLYWFLQR PGQSPQLLIY RMSNLASGVP DR FSGSGSG TAFTLTISRL EAEDVGVYYC MQHLEYPLTF GAGTKLEL |
-Macromolecule #5: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #6: CYCLOHEXYLAMMONIUM ION
| Macromolecule | Name: CYCLOHEXYLAMMONIUM ION / type: ligand / ID: 6 / Number of copies: 1 / Formula: HAI |
|---|---|
| Molecular weight | Theoretical: 100.182 Da |
| Chemical component information | 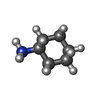 ChemComp-HAI: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 1.875 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: DIFFRACTION / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.0 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.1 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 296951 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller


































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)