+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3724 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The structure of the COPI coat linkage IV | |||||||||
 Map data Map data | The structure of the COPI coat linkage IV | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | COPI / coatomer / coated vesicles / Transport protein | |||||||||
| Function / homology |  Function and homology information Function and homology informationcerebellar Purkinje cell layer maturation / protein localization to cell leading edge / Synthesis of PIPs at the plasma membrane / protein localization to axon / VxPx cargo-targeting to cilium / Synthesis of PIPs at the Golgi membrane / Intra-Golgi traffic / trans-Golgi Network Vesicle Budding / protein localization to Golgi membrane / Golgi localization ...cerebellar Purkinje cell layer maturation / protein localization to cell leading edge / Synthesis of PIPs at the plasma membrane / protein localization to axon / VxPx cargo-targeting to cilium / Synthesis of PIPs at the Golgi membrane / Intra-Golgi traffic / trans-Golgi Network Vesicle Budding / protein localization to Golgi membrane / Golgi localization / COPI-coated vesicle / pancreatic juice secretion / regulation of Golgi organization / organelle membrane contact site / COPI vesicle coat / COPI-dependent Golgi-to-ER retrograde traffic / COPI-mediated anterograde transport / COPI-mediated anterograde transport / Golgi vesicle transport / COPI-dependent Golgi-to-ER retrograde traffic / positive regulation of mitochondrial fusion / organelle transport along microtubule / regulation of fatty acid metabolic process / establishment of Golgi localization / intra-Golgi vesicle-mediated transport / Golgi to plasma membrane transport / retrograde vesicle-mediated transport, Golgi to endoplasmic reticulum / pigmentation / Golgi-associated vesicle / positive regulation of mitochondrial fission / endoplasmic reticulum-Golgi intermediate compartment / protein secretion / endoplasmic reticulum to Golgi vesicle-mediated transport / vesicle-mediated transport / Neutrophil degranulation / adult locomotory behavior / small monomeric GTPase / establishment of localization in cell / protein kinase C binding / macroautophagy / intracellular protein transport / hormone activity / protein transport / growth cone / Golgi membrane / axon / GTPase activity / mRNA binding / neuronal cell body / GTP binding / structural molecule activity / Golgi apparatus / endoplasmic reticulum / extracellular space / nucleoplasm / plasma membrane / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
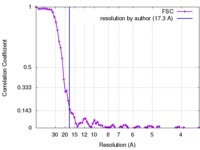
| Method | subtomogram averaging / cryo EM / Resolution: 17.3 Å | |||||||||
 Authors Authors | Dodonova SO / Aderhold P | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: Elife / Year: 2017 Journal: Elife / Year: 2017Title: 9Å structure of the COPI coat reveals that the Arf1 GTPase occupies two contrasting molecular environments. Authors: Svetlana O Dodonova / Patrick Aderhold / Juergen Kopp / Iva Ganeva / Simone Röhling / Wim J H Hagen / Irmgard Sinning / Felix Wieland / John A G Briggs /   Abstract: COPI coated vesicles mediate trafficking within the Golgi apparatus and between the Golgi and the endoplasmic reticulum. Assembly of a COPI coated vesicle is initiated by the small GTPase Arf1 that ...COPI coated vesicles mediate trafficking within the Golgi apparatus and between the Golgi and the endoplasmic reticulum. Assembly of a COPI coated vesicle is initiated by the small GTPase Arf1 that recruits the coatomer complex to the membrane, triggering polymerization and budding. The vesicle uncoats before fusion with a target membrane. Coat components are structurally conserved between COPI and clathrin/adaptor proteins. Using cryo-electron tomography and subtomogram averaging, we determined the structure of the COPI coat assembled on membranes in vitro at 9 Å resolution. We also obtained a 2.57 Å resolution crystal structure of βδ-COP. By combining these structures we built a molecular model of the coat. We additionally determined the coat structure in the presence of ArfGAP proteins that regulate coat dissociation. We found that Arf1 occupies contrasting molecular environments within the coat, leading us to hypothesize that some Arf1 molecules may regulate vesicle assembly while others regulate coat disassembly. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3724.map.gz emd_3724.map.gz | 33.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3724-v30.xml emd-3724-v30.xml emd-3724.xml emd-3724.xml | 27.9 KB 27.9 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3724_fsc.xml emd_3724_fsc.xml | 8.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_3724.png emd_3724.png | 127.2 KB | ||
| Filedesc metadata |  emd-3724.cif.gz emd-3724.cif.gz | 10.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3724 http://ftp.pdbj.org/pub/emdb/structures/EMD-3724 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3724 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3724 | HTTPS FTP |
-Validation report
| Summary document |  emd_3724_validation.pdf.gz emd_3724_validation.pdf.gz | 282.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3724_full_validation.pdf.gz emd_3724_full_validation.pdf.gz | 282.1 KB | Display | |
| Data in XML |  emd_3724_validation.xml.gz emd_3724_validation.xml.gz | 9.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3724 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3724 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3724 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3724 | HTTPS FTP |
-Related structure data
| Related structure data |  5nzvMC  3720C  3721C  3722C  3723C  5mu7C  5nzrC  5nzsC  5nztC  5nzuC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3724.map.gz / Format: CCP4 / Size: 36.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3724.map.gz / Format: CCP4 / Size: 36.3 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
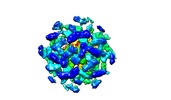
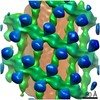
| Annotation | The structure of the COPI coat linkage IV | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.78 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
+Entire : The structure of the COPI coat leaf
+Supramolecule #1: The structure of the COPI coat leaf
+Supramolecule #2: COPI coat complex
+Supramolecule #3: ADP-ribosylation factor 1
+Macromolecule #1: Coatomer subunit alpha
+Macromolecule #2: Coatomer subunit epsilon
+Macromolecule #3: Coatomer subunit beta
+Macromolecule #4: Coatomer subunit beta'
+Macromolecule #5: Coatomer subunit delta
+Macromolecule #6: ADP-ribosylation factor 1
+Macromolecule #7: Coatomer subunit gamma-1
+Macromolecule #8: Coatomer subunit zeta-1
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | subtomogram averaging |
| Aggregation state | 3D array |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 Component:
Details: Protein-A conjugated 10 nm gold was added to the reaction mix in 1:6 volume ratio before plunge-freezing | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Grid | Model: C-flat / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Support film - topology: HOLEY / Pretreatment - Type: GLOW DISCHARGE / Pretreatment - Time: 30 sec. / Pretreatment - Atmosphere: AIR / Details: Protochips C-flat MultiHole 20 mA | ||||||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 85 % / Chamber temperature: 296 K / Instrument: HOMEMADE PLUNGER Details: The sample was applied onto glow-discharged (30 sec, 20 mA) C-flat (Protochips Inc.) multihole grids. The grids were blotted from the back side for 11 seconds at room temperature in a ...Details: The sample was applied onto glow-discharged (30 sec, 20 mA) C-flat (Protochips Inc.) multihole grids. The grids were blotted from the back side for 11 seconds at room temperature in a chamber at 85% humidity and plunge-frozen into liquid ethane using a manual plunger.. | ||||||||||||
| Details | COPI-coated vesicles were produced in vitro by incubating coatomer, Arf1, GTPgS, ARNO and GUVs in a total volume of 40 ul for 30 minutes at 37C |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Energy filter - Name: GIF Quantum LS / Energy filter - Lower energy threshold: 0 eV / Energy filter - Upper energy threshold: 20 eV |
| Details | Tomographic tilt series were acquired with the dose-symmetric tilt-scheme (Hagen et al., J Struct Biol. 2017) |
| Image recording | Film or detector model: GATAN K2 QUANTUM (4k x 4k) / Detector mode: SUPER-RESOLUTION / Digitization - Frames/image: 1-5 / Number grids imaged: 1 / Average electron dose: 2.0 e/Å2 Details: Each of the images in the tilt series was low-pass filtered according to the electron-dose acquired by the sample (Grant and Grigorieff, 2015). |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 5.0 µm / Nominal defocus min: 2.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL / Protocol: RIGID BODY FIT / Target criteria: Cross-correlation coefficient |
|---|---|
| Output model |  PDB-5nzv: |
 Movie
Movie Controller
Controller





















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)