[English] 日本語
 Yorodumi
Yorodumi- EMDB-3712: Human BBsome core complex with subunits BBS1, BBS4, BBS5, BBS8, B... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3712 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
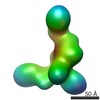
| Title | Human BBsome core complex with subunits BBS1, BBS4, BBS5, BBS8, BBS9, BBS18 | |||||||||
 Map data Map data | Negative Stain Reconstruction:BBsome Complex | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / negative staining / Resolution: 23.0 Å | |||||||||
 Authors Authors | Klink BU / Zent E / Juneja P / Kuhlee A / Raunser S / Wittinghofer A | |||||||||
 Citation Citation |  Journal: Elife / Year: 2017 Journal: Elife / Year: 2017Title: A recombinant BBSome core complex and how it interacts with ciliary cargo. Authors: Björn Udo Klink / Eldar Zent / Puneet Juneja / Anne Kuhlee / Stefan Raunser / Alfred Wittinghofer /  Abstract: Cilia are small, antenna-like structures on the surface of eukaryotic cells that harbor a unique set of sensory proteins, including GPCRs and other membrane proteins. The transport of these proteins ...Cilia are small, antenna-like structures on the surface of eukaryotic cells that harbor a unique set of sensory proteins, including GPCRs and other membrane proteins. The transport of these proteins involves the BBSome, an eight-membered protein complex that is recruited to ciliary membranes by the G-protein Arl6. BBSome malfunction leads to Bardet-Biedl syndrome, a ciliopathy with severe consequences. Short ciliary targeting sequences (CTS) have been identified that trigger the transport of ciliary proteins. However, mechanistic studies that relate ciliary targeting to BBSome binding are missing. Here we used heterologously expressed BBSome subcomplexes to analyze the complex architecture and to investigate the binding of GPCRs and other receptors to the BBSome. A stable heterohexameric complex was identified that binds to GPCRs with interactions that only partially overlap with previously described CTS, indicating a more complex recognition than anticipated. Arl6•GTP does not affect these interactions, suggesting no direct involvement in cargo loading/unloading. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3712.map.gz emd_3712.map.gz | 399.2 KB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3712-v30.xml emd-3712-v30.xml emd-3712.xml emd-3712.xml | 8.8 KB 8.8 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3712_1.png emd_3712_1.png emd_3712_2.png emd_3712_2.png | 29.4 KB 28.8 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3712 http://ftp.pdbj.org/pub/emdb/structures/EMD-3712 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3712 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3712 | HTTPS FTP |
-Validation report
| Summary document |  emd_3712_validation.pdf.gz emd_3712_validation.pdf.gz | 179.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3712_full_validation.pdf.gz emd_3712_full_validation.pdf.gz | 178.5 KB | Display | |
| Data in XML |  emd_3712_validation.xml.gz emd_3712_validation.xml.gz | 5.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3712 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3712 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3712 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3712 | HTTPS FTP |
-Related structure data
| Similar structure data |
|---|
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_3712.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3712.map.gz / Format: CCP4 / Size: 22.2 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Negative Stain Reconstruction:BBsome Complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.89 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : A 6subunit, BBsome complex.
| Entire | Name: A 6subunit, BBsome complex. |
|---|---|
| Components |
|
-Supramolecule #1: A 6subunit, BBsome complex.
| Supramolecule | Name: A 6subunit, BBsome complex. / type: complex / ID: 1 / Parent: 0 Details: Subunits: BBsome subunit 1 BBsome subunit 4 BBsome subunit 5 BBsome subunit 8 BBsome subunit 9 BBsome subunit18 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) / Recombinant cell: High Five cells Trichoplusia ni (cabbage looper) / Recombinant cell: High Five cells |
| Molecular weight | Theoretical: 330 KDa |
-Experimental details
-Structure determination
| Method | negative staining |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 Details: 50 mM Hepes pH 8.0, 150 mM NaCl, 5 mM MgCl2, 10% glycerol and 1 mM TCEP |
|---|---|
| Staining | Type: NEGATIVE / Material: Uranyl formate |
| Grid | Pretreatment - Type: GLOW DISCHARGE Details: Glow discharged 400 mesh copper grids with thin carbon support. |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 1400 |
|---|---|
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number real images: 197 / Average electron dose: 35.0 e/Å2 |
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 3.4 mm / Nominal defocus max: 1.8 µm / Nominal defocus min: 0.9 µm / Nominal magnification: 60000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC |
- Image processing
Image processing
| Final reconstruction | Number classes used: 140 / Applied symmetry - Point group: C1 (asymmetric) / Resolution.type: BY AUTHOR / Resolution: 23.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: SPARX / Number images used: 8339 |
|---|---|
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller


 UCSF Chimera
UCSF Chimera





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















