[English] 日本語
 Yorodumi
Yorodumi- EMDB-36937: post-occluded structure of human ABCB6 W546A mutant (ADP/VO4-bound) -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | post-occluded structure of human ABCB6 W546A mutant (ADP/VO4-bound) | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | ATP-Binding Cassette transporter mitochondrial transporter / post-occluded / ADP/VO4-bound / ABCB6 / MEMBRANE PROTEIN / TRANSPORT PROTEIN | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationDefective ABCB6 causes MCOPCB7 / cellular detoxification of cadmium ion / Mitochondrial ABC transporters / tetrapyrrole metabolic process / ABC-type heme transporter / porphyrin-containing compound metabolic process / tetrapyrrole binding / heme metabolic process / heme transport / porphyrin-containing compound biosynthetic process ...Defective ABCB6 causes MCOPCB7 / cellular detoxification of cadmium ion / Mitochondrial ABC transporters / tetrapyrrole metabolic process / ABC-type heme transporter / porphyrin-containing compound metabolic process / tetrapyrrole binding / heme metabolic process / heme transport / porphyrin-containing compound biosynthetic process / melanosome assembly / heme transmembrane transport / ABC-type heme transporter activity / melanosome membrane / multivesicular body membrane / endolysosome membrane / mitochondrial envelope / vacuolar membrane / skin development / efflux transmembrane transporter activity / intracellular copper ion homeostasis / ABC-type transporter activity / ATP-binding cassette (ABC) transporter complex / brain development / transmembrane transport / early endosome membrane / intracellular iron ion homeostasis / mitochondrial outer membrane / endosome / lysosomal membrane / Golgi membrane / heme binding / endoplasmic reticulum membrane / Golgi apparatus / endoplasmic reticulum / ATP hydrolysis activity / mitochondrion / extracellular exosome / nucleoplasm / ATP binding / plasma membrane / cytosol Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | ||||||||||||
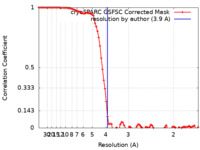
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | ||||||||||||
 Authors Authors | Jin MS / Lee SS / Park JG / Jang E / Choi SH / Kim S / Kim JW | ||||||||||||
| Funding support |  Korea, Republic Of, 3 items Korea, Republic Of, 3 items
| ||||||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2023 Journal: Commun Biol / Year: 2023Title: W546 stacking disruption traps the human porphyrin transporter ABCB6 in an outward-facing transient state. Authors: Sang Soo Lee / Jun Gyou Park / Eunhong Jang / Seung Hun Choi / Subin Kim / Ji Won Kim / Mi Sun Jin /  Abstract: Human ATP-binding cassette transporter subfamily B6 (ABCB6) is a mitochondrial ATP-driven pump that translocates porphyrins from the cytoplasm into mitochondria for heme biosynthesis. Within the ...Human ATP-binding cassette transporter subfamily B6 (ABCB6) is a mitochondrial ATP-driven pump that translocates porphyrins from the cytoplasm into mitochondria for heme biosynthesis. Within the transport pathway, a conserved aromatic residue W546 located in each monomer plays a pivotal role in stabilizing the occluded conformation via π-stacking interactions. Herein, we employed cryo-electron microscopy to investigate the structural consequences of a single W546A mutation in ABCB6, both in detergent micelles and nanodiscs. The results demonstrate that the W546A mutation alters the conformational dynamics of detergent-purified ABCB6, leading to entrapment of the transporter in an outward-facing transient state. However, in the nanodisc system, we observed a direct interaction between the transporter and a phospholipid molecule that compensates for the absence of the W546 residue, thereby facilitating the normal conformational transition of the transporter toward the occluded state following ATP hydrolysis. The findings also reveal that adoption of the outward-facing conformation causes charge repulsion between ABCB6 and the bound substrate, and rearrangement of key interacting residues at the substrate-binding site. Consequently, the affinity for the substrate is significantly reduced, facilitating its release from the transporter. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36937.map.gz emd_36937.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36937-v30.xml emd-36937-v30.xml emd-36937.xml emd-36937.xml | 15.5 KB 15.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36937_fsc.xml emd_36937_fsc.xml | 8.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_36937.png emd_36937.png | 18.7 KB | ||
| Filedesc metadata |  emd-36937.cif.gz emd-36937.cif.gz | 6 KB | ||
| Others |  emd_36937_half_map_1.map.gz emd_36937_half_map_1.map.gz emd_36937_half_map_2.map.gz emd_36937_half_map_2.map.gz | 59.2 MB 59.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36937 http://ftp.pdbj.org/pub/emdb/structures/EMD-36937 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36937 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36937 | HTTPS FTP |
-Validation report
| Summary document |  emd_36937_validation.pdf.gz emd_36937_validation.pdf.gz | 852.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36937_full_validation.pdf.gz emd_36937_full_validation.pdf.gz | 851.7 KB | Display | |
| Data in XML |  emd_36937_validation.xml.gz emd_36937_validation.xml.gz | 16.1 KB | Display | |
| Data in CIF |  emd_36937_validation.cif.gz emd_36937_validation.cif.gz | 20.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36937 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36937 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36937 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36937 | HTTPS FTP |
-Related structure data
| Related structure data |  8k7bMC  8k7cC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36937.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36937.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36937_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_36937_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : ATP-binding cassette sub-family B member 6 - Homo sapiens
| Entire | Name: ATP-binding cassette sub-family B member 6 - Homo sapiens |
|---|---|
| Components |
|
-Supramolecule #1: ATP-binding cassette sub-family B member 6 - Homo sapiens
| Supramolecule | Name: ATP-binding cassette sub-family B member 6 - Homo sapiens type: organelle_or_cellular_component / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: ATP-binding cassette sub-family B member 6
| Macromolecule | Name: ATP-binding cassette sub-family B member 6 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: ABC-type heme transporter |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 66.214156 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: TWRDFGRKLR LLSGYLWPRG SPALQLVVLI CLGLMGLERA LNVLVPIFYR NIVNLLTEKA PWNSLAWTVT SYVFLKFLQG GGTGSTGFV SNLRTFLWIR VQQFTSRRVE LLIFSHLHEL SLRWHLGRRT GEVLRIADRG TSSVTGLLSY LVFNVIPTLA D IIIGIIYF ...String: TWRDFGRKLR LLSGYLWPRG SPALQLVVLI CLGLMGLERA LNVLVPIFYR NIVNLLTEKA PWNSLAWTVT SYVFLKFLQG GGTGSTGFV SNLRTFLWIR VQQFTSRRVE LLIFSHLHEL SLRWHLGRRT GEVLRIADRG TSSVTGLLSY LVFNVIPTLA D IIIGIIYF SMFFNAWFGL IVFLCMSLYL TLTIVVTEWR TKFRRAMNTQ ENATRARAVD SLLNFETVKY YNAESYEVER YR EAIIKYQ GLEWKSSASL VLLNQTQNLV IGLGLLAGSL LCAYFVTEQK LQVGDYVLFG TYIIQLYMPL NAFGTYYRMI QTN FIDMEN MFDLLKEETE VKDLPGAGPL RFQKGRIEFE NVHFSYADGR ETLQDVSFTV MPGQTLALVG PSGAGKSTIL RLLF RFYDI SSGCIRIDGQ DISQVTQASL RSHIGVVPQD TVLFNDTIAD NIRYGRVTAG NDEVEAAAQA AGIHDAIMAF PEGYR TQVG ERGLKLSGGE KQRVAIARTI LKAPGIILLD EATSALDTSN ERAIQASLAK VCANRTTIVV AHRLSTVVNA DQILVI KDG CIVERGRHEA LLSRGGVYAD MWQLQQG UniProtKB: ATP-binding cassette sub-family B member 6 |
-Macromolecule #2: (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY...
| Macromolecule | Name: (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE type: ligand / ID: 2 / Number of copies: 2 / Formula: PEV |
|---|---|
| Molecular weight | Theoretical: 720.012 Da |
| Chemical component information |  ChemComp-PEV: |
-Macromolecule #3: ADP ORTHOVANADATE
| Macromolecule | Name: ADP ORTHOVANADATE / type: ligand / ID: 3 / Number of copies: 2 / Formula: AOV |
|---|---|
| Molecular weight | Theoretical: 544.156 Da |
| Chemical component information | 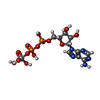 ChemComp-AOV: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 2 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.5 mg/mL |
|---|---|
| Buffer | pH: 7 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K |
- Electron microscopy
Electron microscopy
| Microscope | FEI TALOS ARCTICA |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.4 µm / Nominal defocus min: 0.7000000000000001 µm |
| Experimental equipment |  Model: Talos Arctica / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)