[English] 日本語
 Yorodumi
Yorodumi- EMDB-36755: Structure of human C5a-desArg bound human C5aR1 in complex with Go -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of human C5a-desArg bound human C5aR1 in complex with Go | ||||||||||||
 Map data Map data | |||||||||||||
 Sample Sample |
| ||||||||||||
 Keywords Keywords | GPCR / Cell Signaling / Immune system / SIGNALING PROTEIN-IMMUNE SYSTEM complex | ||||||||||||
| Function / homology |  Function and homology information Function and homology informationcomplement component C5a signaling pathway / presynapse organization / regulation of tau-protein kinase activity / complement component C5a receptor activity / response to peptidoglycan / Terminal pathway of complement / membrane attack complex / sensory perception of chemical stimulus / mu-type opioid receptor binding / Activation of C3 and C5 ...complement component C5a signaling pathway / presynapse organization / regulation of tau-protein kinase activity / complement component C5a receptor activity / response to peptidoglycan / Terminal pathway of complement / membrane attack complex / sensory perception of chemical stimulus / mu-type opioid receptor binding / Activation of C3 and C5 / complement receptor mediated signaling pathway / corticotropin-releasing hormone receptor 1 binding / negative regulation of macrophage chemotaxis / vesicle docking involved in exocytosis / complement activation, alternative pathway / positive regulation of neutrophil chemotaxis / chemokine activity / G protein-coupled dopamine receptor signaling pathway / endopeptidase inhibitor activity / regulation of heart contraction / positive regulation of macrophage chemotaxis / amyloid-beta clearance / positive regulation of vascular endothelial growth factor production / cellular defense response / negative regulation of insulin secretion / complement activation, classical pathway / G protein-coupled serotonin receptor binding / positive regulation of chemokine production / muscle contraction / neutrophil chemotaxis / Peptide ligand-binding receptors / positive regulation of epithelial cell proliferation / secretory granule membrane / Regulation of Complement cascade / locomotory behavior / astrocyte activation / G protein-coupled receptor activity / microglial cell activation / mRNA transcription by RNA polymerase II / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / G-protein activation / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / cognition / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Sensory perception of sweet, bitter, and umami (glutamate) taste / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / photoreceptor disc membrane / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / positive regulation of angiogenesis / sensory perception of taste / ADP signalling through P2Y purinoceptor 1 / G beta:gamma signalling through PI3Kgamma / adenylate cyclase-activating dopamine receptor signaling pathway / chemotaxis / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / cellular response to prostaglandin E stimulus / Inactivation, recovery and regulation of the phototransduction cascade / G-protein beta-subunit binding / heterotrimeric G-protein complex / G alpha (12/13) signalling events / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / apical part of cell / GTPase binding / retina development in camera-type eye / Ca2+ pathway / phospholipase C-activating G protein-coupled receptor signaling pathway / positive regulation of cytosolic calcium ion concentration / cell body / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / basolateral plasma membrane / G alpha (q) signalling events / Ras protein signal transduction / killing of cells of another organism / cell population proliferation / Extra-nuclear estrogen signaling / positive regulation of ERK1 and ERK2 cascade Similarity search - Function | ||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | ||||||||||||
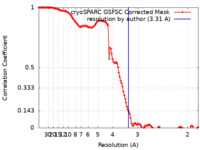
| Method | single particle reconstruction / cryo EM / Resolution: 3.31 Å | ||||||||||||
 Authors Authors | Yadav MK / Yadav R / Maharana J / Sarma P / Banerjee R / Shukla AK / Gati C | ||||||||||||
| Funding support |  United Kingdom, United Kingdom,  India, 3 items India, 3 items
| ||||||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Molecular basis of anaphylatoxin binding, activation, and signaling bias at complement receptors. Authors: Manish K Yadav / Jagannath Maharana / Ravi Yadav / Shirsha Saha / Parishmita Sarma / Chahat Soni / Vinay Singh / Sayantan Saha / Manisankar Ganguly / Xaria X Li / Samanwita Mohapatra / Sudha ...Authors: Manish K Yadav / Jagannath Maharana / Ravi Yadav / Shirsha Saha / Parishmita Sarma / Chahat Soni / Vinay Singh / Sayantan Saha / Manisankar Ganguly / Xaria X Li / Samanwita Mohapatra / Sudha Mishra / Htet A Khant / Mohamed Chami / Trent M Woodruff / Ramanuj Banerjee / Arun K Shukla / Cornelius Gati /     Abstract: The complement system is a critical part of our innate immune response, and the terminal products of this cascade, anaphylatoxins C3a and C5a, exert their physiological and pathophysiological ...The complement system is a critical part of our innate immune response, and the terminal products of this cascade, anaphylatoxins C3a and C5a, exert their physiological and pathophysiological responses primarily via two GPCRs, C3aR and C5aR1. However, the molecular mechanism of ligand recognition, activation, and signaling bias of these receptors remains mostly elusive. Here, we present nine cryo-EM structures of C3aR and C5aR1 activated by their natural and synthetic agonists, which reveal distinct binding pocket topologies of complement anaphylatoxins and provide key insights into receptor activation and transducer coupling. We also uncover the structural basis of a naturally occurring mechanism to dampen the inflammatory response of C5a via proteolytic cleavage of the terminal arginine and the G-protein signaling bias elicited by a peptide agonist of C3aR identified here. In summary, our study elucidates the innerworkings of the complement anaphylatoxin receptors and should facilitate structure-guided drug discovery to target these receptors in a spectrum of disorders. | ||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36755.map.gz emd_36755.map.gz | 167.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36755-v30.xml emd-36755-v30.xml emd-36755.xml emd-36755.xml | 23.2 KB 23.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36755_fsc.xml emd_36755_fsc.xml | 11.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_36755.png emd_36755.png | 83.5 KB | ||
| Filedesc metadata |  emd-36755.cif.gz emd-36755.cif.gz | 6.8 KB | ||
| Others |  emd_36755_half_map_1.map.gz emd_36755_half_map_1.map.gz emd_36755_half_map_2.map.gz emd_36755_half_map_2.map.gz | 165 MB 165 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36755 http://ftp.pdbj.org/pub/emdb/structures/EMD-36755 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36755 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36755 | HTTPS FTP |
-Validation report
| Summary document |  emd_36755_validation.pdf.gz emd_36755_validation.pdf.gz | 817.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36755_full_validation.pdf.gz emd_36755_full_validation.pdf.gz | 817.2 KB | Display | |
| Data in XML |  emd_36755_validation.xml.gz emd_36755_validation.xml.gz | 20 KB | Display | |
| Data in CIF |  emd_36755_validation.cif.gz emd_36755_validation.cif.gz | 25.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36755 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36755 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36755 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36755 | HTTPS FTP |
-Related structure data
| Related structure data |  8jzzMC  8hptC  8hqcC  8i95C  8i97C  8i9aC  8i9lC  8i9sC  8ia2C  8j6dC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36755.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36755.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.92 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_36755_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
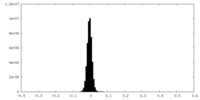
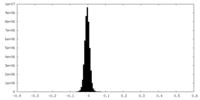
| Density Histograms |
-Half map: #1
| File | emd_36755_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
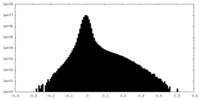
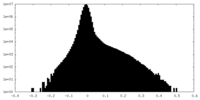
| Density Histograms |
- Sample components
Sample components
-Entire : human C5a-desArg bound human C5aR1 in complex with Go
| Entire | Name: human C5a-desArg bound human C5aR1 in complex with Go |
|---|---|
| Components |
|
-Supramolecule #1: human C5a-desArg bound human C5aR1 in complex with Go
| Supramolecule | Name: human C5a-desArg bound human C5aR1 in complex with Go / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|
-Supramolecule #2: human C5a-desArg bound human C5aR1 in complex with Go
| Supramolecule | Name: human C5a-desArg bound human C5aR1 in complex with Go / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#3, #5-#6 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Supramolecule #3: antibody
| Supramolecule | Name: antibody / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #4 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.534062 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MHHHHHHGSS GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKL IIWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC R FLDDNQIV ...String: MHHHHHHGSS GSELDQLRQE AEQLKNQIRD ARKACADATL SQITNNIDPV GRIQMRTRRT LRGHLAKIYA MHWGTDSRLL VSASQDGKL IIWDSYTTNK VHAIPLRSSW VMTCAYAPSG NYVACGGLDN ICSIYNLKTR EGNVRVSREL AGHTGYLSCC R FLDDNQIV TSSGDTTCAL WDIETGQQTT TFTGHTGDVM SLSLAPDTRL FVSGACDASA KLWDVREGMC RQTFTGHESD IN AICFFPN GNAFATGSDD ATCRLFDLRA DQELMTYSHD NIICGITSVS FSKSGRLLLA GYDDFNCNVW DALKADRAGV LAG HDNRVS CLGVTDDGMA VATGSWDSFL KIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #2: Guanine nucleotide-binding protein G(o) subunit alpha
| Macromolecule | Name: Guanine nucleotide-binding protein G(o) subunit alpha / type: protein_or_peptide / ID: 2 Details: This is a variant of Guanine nucleotide-binding protein G(o) subunit alpha called the mini G(o) alpha Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 27.024762 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGHHHHHHEN LYFQGTLSAE ERAALERSKA IEKNLKEDGI SAAKDVKLLL LGADNSGKST IVKQMKIIHG GSGGSGGTTG IVETHFTFK NLHFRLFDVG GQRSERKKWI HCFEDVTAII FCVDLSDYNR MHESLMLFDS ICNNKFFIDT SIILFLNKKD L FGEKIKKS ...String: MGHHHHHHEN LYFQGTLSAE ERAALERSKA IEKNLKEDGI SAAKDVKLLL LGADNSGKST IVKQMKIIHG GSGGSGGTTG IVETHFTFK NLHFRLFDVG GQRSERKKWI HCFEDVTAII FCVDLSDYNR MHESLMLFDS ICNNKFFIDT SIILFLNKKD L FGEKIKKS PLTICFPEYT GPNTYEDAAA YIQAQFESKN RSPNKEIYCH MTCATDTNNA QVIFDAVTDI IIANNLRGCG LY UniProtKB: Guanine nucleotide-binding protein G(o) subunit alpha, Guanine nucleotide-binding protein G(o) subunit alpha |
-Macromolecule #3: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #4: Antibody fragment ScFv16
| Macromolecule | Name: Antibody fragment ScFv16 / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 26.466486 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS ...String: DVQLVESGGG LVQPGGSRKL SCSASGFAFS SFGMHWVRQA PEKGLEWVAY ISSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSE DTAMYYCVRS IYYYGSSPFD FWGQGTTLTV SSGGGGSGGG GSGGGGSDIV MTQATSSVPV TPGESVSISC R SSKSLLHS NGNTYLYWFL QRPGQSPQLL IYRMSNLASG VPDRFSGSGS GTAFTLTISR LEAEDVGVYY CMQHLEYPLT FG AGTKLEL K |
-Macromolecule #5: C5a anaphylatoxin chemotactic receptor 1
| Macromolecule | Name: C5a anaphylatoxin chemotactic receptor 1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 45.273867 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGKTIIALSY IFCLVFADYK DDDDAANFTP VNGSSGNQSV RLVTSSSLEV LFQGPGSDSF NYTTPDYGHY DDKDTLDLNT PVDKTSNTL RVPDILALVI FAVVFLVGVL GNALVVWVTA FEAKRTINAI WFLNLAVADF LSCLALPILF TSIVQHHHWP F GGAACSIL ...String: MGKTIIALSY IFCLVFADYK DDDDAANFTP VNGSSGNQSV RLVTSSSLEV LFQGPGSDSF NYTTPDYGHY DDKDTLDLNT PVDKTSNTL RVPDILALVI FAVVFLVGVL GNALVVWVTA FEAKRTINAI WFLNLAVADF LSCLALPILF TSIVQHHHWP F GGAACSIL PSLILLNMYA SILLLATISA DRFLLVFKPI WCQNFRGAGL AWIACAVAWG LALLLTIPSF LYRVVREEYF PP KVLCGVD YSHDKRRERA VAIVRLVLGF LWPLLTLTIC YTFILLRTWS RRATRSTKTL KVVVAVVASF FIFWLPYQVT GIM MSFLEP SSPTFLLLKK LDSLCVSFAY INCCINPIIY VVAGQGFQGR LRKSLPSLLR NVLTEESVVR ESKSFTRSTV DTMA QKTQA V UniProtKB: C5a anaphylatoxin chemotactic receptor 1 |
-Macromolecule #6: C5a anaphylatoxin
| Macromolecule | Name: C5a anaphylatoxin / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 8.288676 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TLQKKIEEIA AKYKHSVVKK CCYDGACVNN DETCEQRAAR ISLGPRCIKA FTECCVVASQ LRANISHKDM QLGR UniProtKB: Complement C5 |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | TFS GLACIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON IV (4k x 4k) / Average electron dose: 58.0 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.0 µm / Nominal defocus min: 0.8 µm |
 Movie
Movie Controller
Controller












































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)