+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Dimer of SARS-CoV-2 BA.2 spike and IBT-CoV144(C1 symmetry) | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Spike / nanobody / dimer / local / VIRAL PROTEIN | |||||||||
| Biological species |  | |||||||||
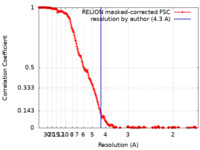
| Method | single particle reconstruction / cryo EM / Resolution: 4.3 Å | |||||||||
 Authors Authors | Yang Y / Zhang CH | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Exploration (Beijing) / Year: 2024 Journal: Exploration (Beijing) / Year: 2024Title: A novel nanobody broadly neutralizes SARS-CoV-2 via induction of spike trimer dimers conformation. Authors: Yang Yang / Junfang Zhang / Shengnan Zhang / Chenhui Zhang / Chenguang Shen / Shuo Song / Yanqun Wang / Yun Peng / Xiaohua Gong / Jun Dai / Chongwei Xie / Tatyana Aleksandrovna Khrustaleva / ...Authors: Yang Yang / Junfang Zhang / Shengnan Zhang / Chenhui Zhang / Chenguang Shen / Shuo Song / Yanqun Wang / Yun Peng / Xiaohua Gong / Jun Dai / Chongwei Xie / Tatyana Aleksandrovna Khrustaleva / Vladislav Victorovich Khrustalev / Yongting Huo / Di Lu / Da Yao / Jincun Zhao / Yingxia Liu / Hongzhou Lu /  Abstract: The ongoing mutations of the SARS-CoV-2 pose serious challenges to the efficacy of the available antiviral drugs, and new drugs with fantastic efficacy are always deserved investigation. Here, a ...The ongoing mutations of the SARS-CoV-2 pose serious challenges to the efficacy of the available antiviral drugs, and new drugs with fantastic efficacy are always deserved investigation. Here, a nanobody called IBT-CoV144 is reported, which exhibits broad neutralizing activity against SARS-CoV-2 by inducing the conformation of spike trimer dimers. IBT-CoV144 was isolated from an immunized alpaca using the RBD of wild-type SARS-CoV-2, and it showed strong cross-reactive binding and neutralizing potency against diverse SARS-CoV-2 variants, including Omicron subvariants. Moreover, the prophylactically and therapeutically intranasal administration of IBT-CoV144 confers fantastic protective efficacy against the challenge of Omicron BA.1 variant in BALB/c mice model. The structure analysis of the complex between spike (S) protein, conducted using Cryo-EM, revealed a special conformation known as the trimer dimers. This conformation is formed by two trimers, with six RBDs in the "up" state and bound by six VHHs. IBT-CoV144 binds to the lateral region of the RBD on the S protein, facilitating the aggregation of S proteins. This aggregation results in steric hindrance, which disrupts the recognition of the virus by ACE2 on host cells. The discovery of IBT-CoV144 will provide valuable insights for the development of advanced therapeutics and the design of next-generation vaccines. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36740.map.gz emd_36740.map.gz | 47.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36740-v30.xml emd-36740-v30.xml emd-36740.xml emd-36740.xml | 14.5 KB 14.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36740_fsc.xml emd_36740_fsc.xml | 22.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_36740.png emd_36740.png | 64.6 KB | ||
| Filedesc metadata |  emd-36740.cif.gz emd-36740.cif.gz | 4.1 KB | ||
| Others |  emd_36740_half_map_1.map.gz emd_36740_half_map_1.map.gz emd_36740_half_map_2.map.gz emd_36740_half_map_2.map.gz | 438.6 MB 438.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36740 http://ftp.pdbj.org/pub/emdb/structures/EMD-36740 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36740 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36740 | HTTPS FTP |
-Validation report
| Summary document |  emd_36740_validation.pdf.gz emd_36740_validation.pdf.gz | 929.5 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36740_full_validation.pdf.gz emd_36740_full_validation.pdf.gz | 929 KB | Display | |
| Data in XML |  emd_36740_validation.xml.gz emd_36740_validation.xml.gz | 28.5 KB | Display | |
| Data in CIF |  emd_36740_validation.cif.gz emd_36740_validation.cif.gz | 38 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36740 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36740 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36740 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36740 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36740.map.gz / Format: CCP4 / Size: 567.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36740.map.gz / Format: CCP4 / Size: 567.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.842 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_36740_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
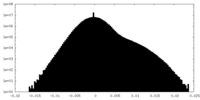
| Projections & Slices |
| ||||||||||||
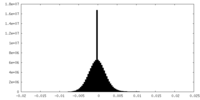
| Density Histograms |
-Half map: #2
| File | emd_36740_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
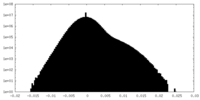
| Projections & Slices |
| ||||||||||||
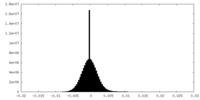
| Density Histograms |
- Sample components
Sample components
-Entire : complex of RBD domain of SARS-CoV-2 spike protein and IBT-CoV144
| Entire | Name: complex of RBD domain of SARS-CoV-2 spike protein and IBT-CoV144 |
|---|---|
| Components |
|
-Supramolecule #1: complex of RBD domain of SARS-CoV-2 spike protein and IBT-CoV144
| Supramolecule | Name: complex of RBD domain of SARS-CoV-2 spike protein and IBT-CoV144 type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL |
|---|---|
| Buffer | pH: 7.4 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Chamber temperature: 277 K / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Temperature | Min: 93.0 K / Max: 93.0 K |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 70.0 µm / Calibrated defocus max: 2.5 µm / Calibrated defocus min: 1.5 µm / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)