+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | membrane proteins | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | enzyme / acetylation / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationheparan-alpha-glucosaminide N-acetyltransferase / heparan-alpha-glucosaminide N-acetyltransferase activity / MPS IIIC - Sanfilippo syndrome C / heparan sulfate proteoglycan catabolic process / HS-GAG degradation / lysosomal transport / acyltransferase activity / protein complex oligomerization / tertiary granule membrane / specific granule membrane ...heparan-alpha-glucosaminide N-acetyltransferase / heparan-alpha-glucosaminide N-acetyltransferase activity / MPS IIIC - Sanfilippo syndrome C / heparan sulfate proteoglycan catabolic process / HS-GAG degradation / lysosomal transport / acyltransferase activity / protein complex oligomerization / tertiary granule membrane / specific granule membrane / lysosomal lumen / lysosomal membrane / Neutrophil degranulation / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
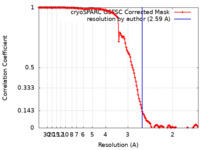
| Method | single particle reconstruction / cryo EM / Resolution: 2.59 Å | |||||||||
 Authors Authors | Yu J / Ge JP / Xu RS | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Struct Mol Biol / Year: 2024 Journal: Nat Struct Mol Biol / Year: 2024Title: Structure and mechanism of lysosome transmembrane acetylation by HGSNAT. Authors: Ruisheng Xu / Yingjie Ning / Fandong Ren / Chenxia Gu / Zhengjiang Zhu / Xuefang Pan / Alexey V Pshezhetsky / Jingpeng Ge / Jie Yu /   Abstract: Lysosomal transmembrane acetylation of heparan sulfates (HS) is catalyzed by HS acetyl-CoA:α-glucosaminide N-acetyltransferase (HGSNAT), whose dysfunction leads to lysosomal storage diseases. The ...Lysosomal transmembrane acetylation of heparan sulfates (HS) is catalyzed by HS acetyl-CoA:α-glucosaminide N-acetyltransferase (HGSNAT), whose dysfunction leads to lysosomal storage diseases. The mechanism by which HGSNAT, the sole non-hydrolase enzyme in HS degradation, brings cytosolic acetyl-coenzyme A (Ac-CoA) and lysosomal HS together for N-acyltransferase reactions remains unclear. Here, we present cryogenic-electron microscopy structures of HGSNAT alone, complexed with Ac-CoA and with acetylated products. These structures explain that Ac-CoA binding from the cytosolic side causes dimeric HGSNAT to form a transmembrane tunnel. Within this tunnel, catalytic histidine and asparagine approach the lumen and instigate the transfer of the acetyl group from Ac-CoA to the glucosamine group of HS. Our study unveils a transmembrane acetylation mechanism that may help advance therapeutic strategies targeting lysosomal storage diseases. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36387.map.gz emd_36387.map.gz | 230 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36387-v30.xml emd-36387-v30.xml emd-36387.xml emd-36387.xml | 16 KB 16 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_36387_fsc.xml emd_36387_fsc.xml | 13.2 KB | Display |  FSC data file FSC data file |
| Images |  emd_36387.png emd_36387.png | 92.7 KB | ||
| Filedesc metadata |  emd-36387.cif.gz emd-36387.cif.gz | 5.9 KB | ||
| Others |  emd_36387_half_map_1.map.gz emd_36387_half_map_1.map.gz emd_36387_half_map_2.map.gz emd_36387_half_map_2.map.gz | 226.5 MB 226.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36387 http://ftp.pdbj.org/pub/emdb/structures/EMD-36387 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36387 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36387 | HTTPS FTP |
-Validation report
| Summary document |  emd_36387_validation.pdf.gz emd_36387_validation.pdf.gz | 797.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_36387_full_validation.pdf.gz emd_36387_full_validation.pdf.gz | 796.7 KB | Display | |
| Data in XML |  emd_36387_validation.xml.gz emd_36387_validation.xml.gz | 21.7 KB | Display | |
| Data in CIF |  emd_36387_validation.cif.gz emd_36387_validation.cif.gz | 27.6 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36387 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36387 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36387 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-36387 | HTTPS FTP |
-Related structure data
| Related structure data |  8jl3MC  8jkvC  8jl1C  8w4aC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_36387.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36387.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.83 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_36387_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_36387_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
+Entire : HGSNAT
+Supramolecule #1: HGSNAT
+Macromolecule #1: Heparan-alpha-glucosaminide N-acetyltransferase
+Macromolecule #3: 2-acetamido-2-deoxy-beta-D-glucopyranose
+Macromolecule #4: ACETYL COENZYME *A
+Macromolecule #5: DODECANE
+Macromolecule #6: N-OCTANE
+Macromolecule #7: TETRADECANE
+Macromolecule #8: CHOLESTEROL
+Macromolecule #9: DECANE
+Macromolecule #10: HEXADECANE
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 0.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)