[English] 日本語
 Yorodumi
Yorodumi- EMDB-36145: Neurokinin B bound to active human neurokinin 3 receptor in compl... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
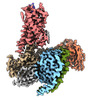
| Title | Neurokinin B bound to active human neurokinin 3 receptor in complex with Gq | |||||||||
 Map data Map data | Neurokinin B bound to active human neurokinin 3 receptor in complex with Gq | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | G protein coupled receptor / Neurokinin / Cryo-EM / Peptide Agonists / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationTachykinin receptors bind tachykinins / tachykinin receptor signaling pathway / positive regulation of blood pressure / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 ...Tachykinin receptors bind tachykinins / tachykinin receptor signaling pathway / positive regulation of blood pressure / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Ca2+ pathway / G alpha (z) signalling events / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / G alpha (q) signalling events / Thrombin signalling through proteinase activated receptors (PARs) / G alpha (i) signalling events / alkylglycerophosphoethanolamine phosphodiesterase activity / Activation of G protein gated Potassium channels / G-protein activation / G beta:gamma signalling through PI3Kgamma / Prostacyclin signalling through prostacyclin receptor / G beta:gamma signalling through PLC beta / ADP signalling through P2Y purinoceptor 1 / Thromboxane signalling through TP receptor / Presynaptic function of Kainate receptors / G beta:gamma signalling through CDC42 / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Glucagon-type ligand receptors / G alpha (12/13) signalling events / G beta:gamma signalling through BTK / ADP signalling through P2Y purinoceptor 12 / Adrenaline,noradrenaline inhibits insulin secretion / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Thrombin signalling through proteinase activated receptors (PARs) / Ca2+ pathway / Extra-nuclear estrogen signaling / G alpha (z) signalling events / G alpha (s) signalling events / G alpha (q) signalling events / photoreceptor outer segment membrane / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (i) signalling events / spectrin binding / Vasopressin regulates renal water homeostasis via Aquaporins / photoreceptor outer segment / neuropeptide signaling pathway / cardiac muscle cell apoptotic process / photoreceptor inner segment / female pregnancy / cellular response to catecholamine stimulus / sensory perception of taste / adenylate cyclase-activating dopamine receptor signaling pathway / cellular response to prostaglandin E stimulus / G-protein beta-subunit binding / heterotrimeric G-protein complex / signaling receptor complex adaptor activity / GTPase binding / retina development in camera-type eye / phospholipase C-activating G protein-coupled receptor signaling pathway / positive regulation of cytosolic calcium ion concentration / cellular response to hypoxia / cell body / G alpha (q) signalling events / cell population proliferation / G protein-coupled receptor signaling pathway / signaling receptor binding / GTPase activity / dendrite / synapse / protein-containing complex binding / extracellular space / extracellular region / membrane / plasma membrane / cytoplasm Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /    | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||
 Authors Authors | Sun WJ / Yang F / Zhang HH / Yuan QN / Yin WC / Shi P / Eric X / Tian CL | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell Discov / Year: 2023 Journal: Cell Discov / Year: 2023Title: Structural insights into neurokinin 3 receptor activation by endogenous and analogue peptide agonists. Authors: Wenjing Sun / Fan Yang / Huanhuan Zhang / Qingning Yuan / Shenglong Ling / Yuanxia Wang / Pei Lv / Zelin Li / Yifan Luo / Dongsheng Liu / Wanchao Yin / Pan Shi / H Eric Xu / Changlin Tian /  Abstract: Neurokinin 3 receptor (NK3R) is a tachykinin receptor essential for the hypothalamic-pituitary-gonadal axis. The endogenous peptide agonist neurokinin B (NKB) preferentially activates NK3R, while ...Neurokinin 3 receptor (NK3R) is a tachykinin receptor essential for the hypothalamic-pituitary-gonadal axis. The endogenous peptide agonist neurokinin B (NKB) preferentially activates NK3R, while substance P (SP) binds preferentially to NK1R. In addition, the SP analogue senktide more potently activates NK3R than NKB and SP. However, the mechanisms of preferential binding of peptide and NK3R activation remain elusive. Herein, we determined the cryogenic electron microscopy (cryo-EM) structures of the NK3R-G complex bound to NKB, SP and senktide. The three NK3R-G/peptide complexes utilize a class of noncanonical receptor activation mechanisms. Combining the structural analysis and functional assay illustrated that the consensus C-termini of the three peptide agonists share a conserved binding mode to NK3R, while the divergent N-termini of the peptides confer the preferential binding of the agonist to NK3R. In addition, the specific interactions between the N-terminus of senktide and the N-terminus and extracellular loops (ECL2 and ECL3) of NK3R lead to the improved activation displayed by senktide compared to SP and NKB. These findings pave the way to understand tachykinin receptor subtype selectivity and provide ideas to rationally develop drugs targeting NK3R. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_36145.map.gz emd_36145.map.gz | 28.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-36145-v30.xml emd-36145-v30.xml emd-36145.xml emd-36145.xml | 21.6 KB 21.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_36145.png emd_36145.png | 159.6 KB | ||
| Filedesc metadata |  emd-36145.cif.gz emd-36145.cif.gz | 7.1 KB | ||
| Others |  emd_36145_half_map_1.map.gz emd_36145_half_map_1.map.gz emd_36145_half_map_2.map.gz emd_36145_half_map_2.map.gz | 28.2 MB 28.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-36145 http://ftp.pdbj.org/pub/emdb/structures/EMD-36145 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36145 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-36145 | HTTPS FTP |
-Related structure data
| Related structure data |  8jbgMC  8jbfC  8jbhC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_36145.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_36145.map.gz / Format: CCP4 / Size: 30.5 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Neurokinin B bound to active human neurokinin 3 receptor in complex with Gq | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Neurokinin B bound to active human neurokinin 3...
| File | emd_36145_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Neurokinin B bound to active human neurokinin 3 receptor in complex with Gq | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Neurokinin B bound to active human neurokinin 3...
| File | emd_36145_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Neurokinin B bound to active human neurokinin 3 receptor in complex with Gq | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Neurokinin B bound to active human neurokinin 3 receptor in compl...
| Entire | Name: Neurokinin B bound to active human neurokinin 3 receptor in complex with Gq heterotrimer |
|---|---|
| Components |
|
-Supramolecule #1: Neurokinin B bound to active human neurokinin 3 receptor in compl...
| Supramolecule | Name: Neurokinin B bound to active human neurokinin 3 receptor in complex with Gq heterotrimer type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Macromolecule #1: Neuromedin-K receptor (NK3R-pFastbac1)
| Macromolecule | Name: Neuromedin-K receptor (NK3R-pFastbac1) / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 158.089531 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTIIALSYI FCLVFAADLE DNWETLNDNL KVIEKADNAA QVKDALTKMR AAALDAQKAT PPKLEDKSPD SPEMKDFRHG FDILVGQID DALKLANEGK VKEAQAAAEQ LKTTRNAYIQ KYLMATLPAA ETWIDGGGGV GADAVNLTAS LAAGAATGAV E TGWLQLLD ...String: MKTIIALSYI FCLVFAADLE DNWETLNDNL KVIEKADNAA QVKDALTKMR AAALDAQKAT PPKLEDKSPD SPEMKDFRHG FDILVGQID DALKLANEGK VKEAQAAAEQ LKTTRNAYIQ KYLMATLPAA ETWIDGGGGV GADAVNLTAS LAAGAATGAV E TGWLQLLD QAGNLSSSPS ALGLPVASPA PSQPWANLTN QFVQPSWRIA LWSLAYGVVV AVAVLGNLIV IWIILAHKRM RT VTNYFLV NLAFSDASMA AFNTLVNFIY ALHSEWYFGA NYCRFQNFFP ITAVFASIYS MTAIAVDRYM AIIDPLKPRL SAT ATKIVI GSIWILAFLL AFPQCLYSKT KVMPGRTLCF VQWPEGPKQH FTYHIIVIIL VYCFPLLIMG ITYTIVGITL WGGE IPGDT CDKYHEQLKA KRKVVKMMII VVMTFAICWL PYHIYFILTA IYQQLNRWKY IQQVYLASFW LAMSSTMYNP IIYCC LNKR FRAGFKRAFR WCPFIKVSSY DELELKTTRF HPVFTLEDFV GDWEQTAAYN LDQVLEQGGV SSLLQNLAVS VTPIQR IVR SGENALKIDI HVIIPYEGLS ADQMAQIEEV FKVVYPVDDH HFKVILPYGT LVIDGVTPNM LNYFGRPYEG IAVFDGK KI TVTGTLWNGN KIIDERLITP DGSMLFRVTI NSGGSENLYF QGGSAGSAAK IEEGKLVIWI NGDKGYNGLA EVGKKFEK D TGIKVTVEHP DKLEEKFPQV AATGDGPDII FWAHDRFGGY AQSGLLAEIT PDKAFQDKLY PFTWDAVRYN GKLIAYPIA VEALSLIYNK DLLPNPPKTW EEIPALDKEL KAKGKSALMF NLQEPYFTWP LIAADGGYAF KYENGKYDIK DVGVDNAGAK AGLTFLVDL IKNKHMNADT DYSIAEAAFN KGETAMTING PWAWSNIDTS KVNYGVTVLP TFKGQPSKPF VGVLSAGINA A SPNKELAK EFLENYLLTD EGLEAVNKDK PLGAVALKSY EEELAKDPRI AATMENAQKG EIMPNIPQMS AFWYAVRTAV IN AASGRQT VDEALKDAQT AKIEEGKLVI WINGDKGYNG LAEVGKKFEK DTGIKVTVEH PDKLEEKFPQ VAATGDGPDI IFW AHDRFG GYAQSGLLAE ITPDKAFQDK LYPFTWDAVR YNGKLIAYPI AVEALSLIYN KDLLPNPPKT WEEIPALDKE LKAK GKSAL MFNLQEPYFT WPLIAADGGY AFKYENGKYD IKDVGVDNAG AKAGLTFLVD LIKNKHMNAD TDYSIAEAAF NKGET AMTI NGPWAWSNID TSKVNYGVTV LPTFKGQPSK PFVGVLSAGI NAASPNKELA KEFLENYLLT DEGLEAVNKD KPLGAV ALK SYEEELAKDP RIAATMENAQ KGEIMPNIPQ MSAFWYAVRT AVINAASGRQ TVDEALKDAQ TRITK |
-Macromolecule #2: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 41.055867 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: HMHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI ...String: HMHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI VTSSGDTTCA LWDIETGQQT TTFTGHTGDV MSLSLAPDTR LFVSGACDAS AKLWDVREGM CRQTFTGHES DI NAICFFP NGNAFATGSD DATCRLFDLR ADQELMTYSH DNIICGITSV SFSKSGRLLL AGYDDFNCNV WDALKADRAG VLA GHDNRV SCLGVTDDGM AVATGSWDSF LKIWNGSSGG GGSGGGGSSG VSGWRLFKKI S UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #3: ScFv16 nanobody
| Macromolecule | Name: ScFv16 nanobody / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 32.708473 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MLLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFAVQ LVESGGGLVQ PGGSRKLSCS ASGFAFSSFG MHWVRQAPEK GLEWVAYIS SGSGTIYYAD TVKGRFTISR DDPKNTLFLQ MTSLRSEDTA MYYCVRSIYY YGSSPFDFWG QGTTLTVSAG G GGSGGGGS ...String: MLLVNQSHQG FNKEHTSKMV SAIVLYVLLA AAAHSAFAVQ LVESGGGLVQ PGGSRKLSCS ASGFAFSSFG MHWVRQAPEK GLEWVAYIS SGSGTIYYAD TVKGRFTISR DDPKNTLFLQ MTSLRSEDTA MYYCVRSIYY YGSSPFDFWG QGTTLTVSAG G GGSGGGGS GGGGSSDIVM TQATSSVPVT PGESVSISCR SSKSLLHSNG NTYLYWFLQR PGQSPQLLIY RMSNLASGVP DR FSGSGSG TAFTLTISRL EAEDVGVYYC MQHLEYPLTF GAGTKLELVD ENLYFQGASH HHHHHHH |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-Macromolecule #5: Neurokinin-B
| Macromolecule | Name: Neurokinin-B / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 1.212417 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: DMHDFFVGLM UniProtKB: Tachykinin-3 |
-Macromolecule #6: Guanine nucleotide-binding protein Gq (G324)
| Macromolecule | Name: Guanine nucleotide-binding protein Gq (G324) / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 41.694289 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGCTLSAEDK AAVERSKMIE KQLQKDKQVY RRTLRLLLLG ADNSGKSTIV KQMRIYHVNG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMIE KQLQKDKQVY RRTLRLLLLG ADNSGKSTIV KQMRIYHVNG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTSGIFETK FQVDKVNFHM FDVGAQRDER RKWIQCFNDV TAIIFVVDSS DYNRLQEALN DF DSIWNNR WLRTISVILF LNKQDLLAEK VLAGKSKIED YFPEFARYTT PEDATPEPGE DPRVTRAKYF IRKEFVDIST ASG DGRHIC YPHFTCAVDT ENARRIFNDC KDIILQMNLR EYNLV |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.0 mg/mL |
|---|---|
| Buffer | pH: 7.5 |
| Vitrification | Cryogen name: ETHANE-PROPANE / Chamber humidity: 100 % |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: DIFFRACTION / Cs: 2.7 mm / Nominal defocus max: 2.2 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: PDB ENTRY PDB model - PDB ID: |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 907878 |
| Initial angle assignment | Type: ANGULAR RECONSTITUTION |
| Final angle assignment | Type: ANGULAR RECONSTITUTION |
 Movie
Movie Controller
Controller















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)