+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of Full-Length AsfvPrimPol in Complex-Form | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | polymerase / primase / PrimPol / Helicase / DNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationhydrolase activity, acting on acid anhydrides / helicase activity / DNA replication / ATP binding / metal ion binding Similarity search - Function | |||||||||
| Biological species |  African swine fever virus BA71V / synthetic construct (others) African swine fever virus BA71V / synthetic construct (others) | |||||||||
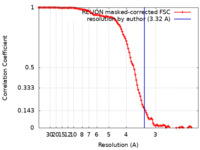
| Method | single particle reconstruction / cryo EM / Resolution: 3.32 Å | |||||||||
 Authors Authors | Shao ZW / Su SC / Gan JH | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nucleic Acids Res / Year: 2023 Journal: Nucleic Acids Res / Year: 2023Title: Structures and implications of the C962R protein of African swine fever virus. Authors: Zhiwei Shao / Shichen Su / Jie Yang / Weizhen Zhang / Yanqing Gao / Xin Zhao / Yixi Zhang / Qiyuan Shao / Chulei Cao / Huili Li / Hehua Liu / Jinru Zhang / Jinzhong Lin / Jinbiao Ma / Jianhua Gan /  Abstract: African swine fever virus (ASFV) is highly contagious and can cause lethal disease in pigs. Although it has been extensively studied in the past, no vaccine or other useful treatment against ASFV is ...African swine fever virus (ASFV) is highly contagious and can cause lethal disease in pigs. Although it has been extensively studied in the past, no vaccine or other useful treatment against ASFV is available. The genome of ASFV encodes more than 170 proteins, but the structures and functions for the majority of the proteins remain elusive, which hindered our understanding on the life cycle of ASFV and the development of ASFV-specific inhibitors. Here, we report the structural and biochemical studies of the highly conserved C962R protein of ASFV, showing that C962R is a multidomain protein. The N-terminal AEP domain is responsible for the DNA polymerization activity, whereas the DNA unwinding activity is catalyzed by the central SF3 helicase domain. The middle PriCT2 and D5_N domains and the C-terminal Tail domain all contribute to the DNA unwinding activity of C962R. C962R preferentially works on forked DNA, and likely functions in Base-excision repair (BER) or other repair pathway in ASFV. Although it is not essential for the replication of ASFV, C962R can serve as a model and provide mechanistic insight into the replicative primase proteins from many other species, such as nitratiruptor phage NrS-1, vaccinia virus (VACV) and other viruses. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35671.map.gz emd_35671.map.gz | 12.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35671-v30.xml emd-35671-v30.xml emd-35671.xml emd-35671.xml | 17.4 KB 17.4 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35671_fsc.xml emd_35671_fsc.xml | 11.4 KB | Display |  FSC data file FSC data file |
| Images |  emd_35671.png emd_35671.png | 135 KB | ||
| Filedesc metadata |  emd-35671.cif.gz emd-35671.cif.gz | 6.4 KB | ||
| Others |  emd_35671_half_map_1.map.gz emd_35671_half_map_1.map.gz emd_35671_half_map_2.map.gz emd_35671_half_map_2.map.gz | 98.2 MB 98.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35671 http://ftp.pdbj.org/pub/emdb/structures/EMD-35671 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35671 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35671 | HTTPS FTP |
-Validation report
| Summary document |  emd_35671_validation.pdf.gz emd_35671_validation.pdf.gz | 721.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35671_full_validation.pdf.gz emd_35671_full_validation.pdf.gz | 721.4 KB | Display | |
| Data in XML |  emd_35671_validation.xml.gz emd_35671_validation.xml.gz | 18.4 KB | Display | |
| Data in CIF |  emd_35671_validation.cif.gz emd_35671_validation.cif.gz | 24.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35671 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35671 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35671 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35671 | HTTPS FTP |
-Related structure data
| Related structure data |  8iqiMC  8iqbC  8iqcC  8iqdC  8iqhC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35671.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35671.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.1 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35671_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_35671_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Full-Length AsfvPrimPol in complex with DNA and AMPPNP
| Entire | Name: Full-Length AsfvPrimPol in complex with DNA and AMPPNP |
|---|---|
| Components |
|
-Supramolecule #1: Full-Length AsfvPrimPol in complex with DNA and AMPPNP
| Supramolecule | Name: Full-Length AsfvPrimPol in complex with DNA and AMPPNP type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 / Details: ASFV ORF C962R |
|---|---|
| Molecular weight | Theoretical: 660 KDa |
-Supramolecule #2: Full-Length AsfvPrimPol
| Supramolecule | Name: Full-Length AsfvPrimPol / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  African swine fever virus BA71V African swine fever virus BA71V |
-Supramolecule #3: DNA
| Supramolecule | Name: DNA / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2 |
|---|
-Macromolecule #1: Putative primase C962R
| Macromolecule | Name: Putative primase C962R / type: protein_or_peptide / ID: 1 / Number of copies: 6 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  African swine fever virus BA71V African swine fever virus BA71V |
| Molecular weight | Theoretical: 111.706273 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GSMREESWEE HDTIQLTAQR KYLAEVQALE TLLARELSVF LTEPGSKKTN IINRITGKTY ALPSTELLRF YEHLEQCRKQ GALMYFLER QGTYSGLMLD YDLKLNTNAA PSLESSVLSR LCHRIFVHIK NSSVLPEGSH KIHFFFTLKP EAVQGKYGFH V LIPGLKMA ...String: GSMREESWEE HDTIQLTAQR KYLAEVQALE TLLARELSVF LTEPGSKKTN IINRITGKTY ALPSTELLRF YEHLEQCRKQ GALMYFLER QGTYSGLMLD YDLKLNTNAA PSLESSVLSR LCHRIFVHIK NSSVLPEGSH KIHFFFTLKP EAVQGKYGFH V LIPGLKMA ASTKKSIIAS LQHDATVQKI LHEQGVANPE SCLDPHSASV PSLLYGSSKL NHRPYQLKTG FELVFDSSDP DY IPIHQIK NIESYNLVSE LSLTNEQGSL VRPVYCAADI AAEKEEEIPA DDHSLSILML HDPEARYLHK ILNLLPPEYY VEY PLWSNV VFALANTSAN YRPLAEWFSQ KCPEKWNTGG KEKLEKLWND ASRHTEKKIT KRSIMYWAHK HAPQQYKEIV EQGY FSILA EYVYSYNGTL EHYMIAKVIY AMMGNKFVVD VDSNGKYVWF EFVLPGQPMN QGEIWKWRKE VNPDELHIYI SENFS RVMD RITEHIKYHL SQPHETNILN YYKKLLKAFE RSKSKIFNDS FKKGVIRQAE FLFRQRSFIQ TLDTNPYLLG VGNGVL SIE TIPAKLINHF HEHPIHQYTH ICYEPFNPEN PWTKLLLNAL QDIIPELDAR LWIMFYLSTA IFRGLKEALM LLWLGGG CN GKTFLMRLVA MVLGDHYASK LNISLLTSYR ETAEKPNSAF MRLKGRGYGY FEETNKSEIL NTSRLKEMVN PGDVTARE L NQKQESFQMT ATMVAASNYN FIIDTTDHGT WRRLRHYRSK VKFCHNPDPN NSYEKKEDPR FIHEYIMDPN CQNAFFSIL VYFWEKLQKE YNGQIKKVFC PTIESETEAY RKSQDTLHRF ITERVVESPS AETVYNLSEV VTAYAEWYNA NINVKRHIAL ELSQELENS VLEKYLQWSP NKTRILKGCR ILHKFETLQP GESYIGVSST GTLLNTPICE PKNKWWEWSP NPSAPPEKEA S APTP UniProtKB: Putative primase C962R |
-Macromolecule #2: DNA (32-MER)
| Macromolecule | Name: DNA (32-MER) / type: dna / ID: 2 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Theoretical: 9.689213 KDa |
| Sequence | String: (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT)(DT) (DT)(DT)(DT) |
-Macromolecule #3: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER
| Macromolecule | Name: PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER / type: ligand / ID: 3 / Number of copies: 6 / Formula: ANP |
|---|---|
| Molecular weight | Theoretical: 506.196 Da |
| Chemical component information |  ChemComp-ANP: |
-Macromolecule #4: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 4 / Number of copies: 4 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.2 mg/mL |
|---|---|
| Buffer | pH: 8 |
| Grid | Model: Quantifoil R1.2/1.3 / Material: COPPER / Mesh: 300 |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 0.01 mm / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.5 µm / Nominal magnification: 64000 |
| Sample stage | Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller









 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)