[English] 日本語
 Yorodumi
Yorodumi- EMDB-35640: Cryo-EM structure of heme transporter CydDC from Mycobacterium sm... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of heme transporter CydDC from Mycobacterium smegmatis in the inward facing apo state | ||||||||||||||||||
 Map data Map data | |||||||||||||||||||
 Sample Sample |
| ||||||||||||||||||
 Keywords Keywords | ABC transporter / Heme / TRANSPORT PROTEIN | ||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsulfate-transporting ATPase / cysteine transport / ABC-type transporter activity / ATP hydrolysis activity / ATP binding / membrane Similarity search - Function | ||||||||||||||||||
| Biological species |  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) | ||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.5 Å | ||||||||||||||||||
 Authors Authors | Zhu C / Li J | ||||||||||||||||||
| Funding support |  China, 5 items China, 5 items
| ||||||||||||||||||
 Citation Citation |  Journal: Protein Cell / Year: 2023 Journal: Protein Cell / Year: 2023Title: Cryo-EM structures of a prokaryotic heme transporter CydDC. Authors: Chen Zhu / Yanfeng Shi / Jing Yu / Wenhao Zhao / Lingqiao Li / Jingxi Liang / Xiaolin Yang / Bing Zhang / Yao Zhao / Yan Gao / Xiaobo Chen / Xiuna Yang / Lu Zhang / Luke W Guddat / Lei Liu / ...Authors: Chen Zhu / Yanfeng Shi / Jing Yu / Wenhao Zhao / Lingqiao Li / Jingxi Liang / Xiaolin Yang / Bing Zhang / Yao Zhao / Yan Gao / Xiaobo Chen / Xiuna Yang / Lu Zhang / Luke W Guddat / Lei Liu / Haitao Yang / Zihe Rao / Jun Li /   | ||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35640.map.gz emd_35640.map.gz | 117.9 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35640-v30.xml emd-35640-v30.xml emd-35640.xml emd-35640.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35640.png emd_35640.png | 62.4 KB | ||
| Filedesc metadata |  emd-35640.cif.gz emd-35640.cif.gz | 5.9 KB | ||
| Others |  emd_35640_half_map_1.map.gz emd_35640_half_map_1.map.gz emd_35640_half_map_2.map.gz emd_35640_half_map_2.map.gz | 116.1 MB 116.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35640 http://ftp.pdbj.org/pub/emdb/structures/EMD-35640 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35640 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35640 | HTTPS FTP |
-Validation report
| Summary document |  emd_35640_validation.pdf.gz emd_35640_validation.pdf.gz | 1.1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35640_full_validation.pdf.gz emd_35640_full_validation.pdf.gz | 1.1 MB | Display | |
| Data in XML |  emd_35640_validation.xml.gz emd_35640_validation.xml.gz | 14 KB | Display | |
| Data in CIF |  emd_35640_validation.cif.gz emd_35640_validation.cif.gz | 16.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35640 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35640 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35640 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35640 | HTTPS FTP |
-Related structure data
| Related structure data |  8ipqMC  8iprC  8ipsC  8iptC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35640.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35640.map.gz / Format: CCP4 / Size: 125 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
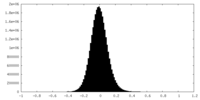
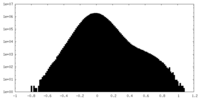
-Half map: #2
| File | emd_35640_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
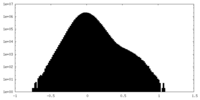
| Projections & Slices |
| ||||||||||||
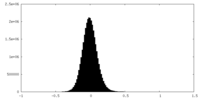
| Density Histograms |
-Half map: #1
| File | emd_35640_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : CydDC
| Entire | Name: CydDC |
|---|---|
| Components |
|
-Supramolecule #1: CydDC
| Supramolecule | Name: CydDC / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) |
-Macromolecule #1: Transmembrane ATP-binding protein ABC transporter cydD
| Macromolecule | Name: Transmembrane ATP-binding protein ABC transporter cydD type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO / EC number: sulfate-transporting ATPase |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) |
| Molecular weight | Theoretical: 53.259984 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) |
| Sequence | String: SYFQSNVVIA GCTIASAVVL AHIVAGIITN PATALGGETD WAPGLVALAV LWSVRVVAQW FQGRLSQRGA TAVIGELSRQ VLSSVTTSS PRRLAADRDS AAAVVTRGLD GLRPYFTGYL PAVVLAGILT PAALVVMAAY DWQAAAIVVI ALPLIPIFMV L IGLLTAER ...String: SYFQSNVVIA GCTIASAVVL AHIVAGIITN PATALGGETD WAPGLVALAV LWSVRVVAQW FQGRLSQRGA TAVIGELSRQ VLSSVTTSS PRRLAADRDS AAAVVTRGLD GLRPYFTGYL PAVVLAGILT PAALVVMAAY DWQAAAIVVI ALPLIPIFMV L IGLLTAER SAAALTAMTT LQGRMLDLIA GIPTLRAVGR AGGSVQRIAE LSASHRRSTM ATLRISFLSA LVLELLATLG VA LVAVSVG LRLVFGDMTL AAGLTALLLA PEVFWPLRRV GAAFHAAQDG KTAAEQALRL CAEPHPPTGH EVVPAGAPVI EVP ALKAVM EPGRVTVLTG PNGVGKSTLL QAILGLQESP CGPILVAGVE VGALDRSAWW GRLAWMPHRP VLVPGTVREN LELL GPVPG LDEVCRSVGF DEVLGELPDG SETPLGRGGV GLSLGQRQRL GLVRALGAPA DVLLLDEPTA HLDGALEDRV LAAIV ARAR AGATVVMVGH RAPVLAAADH VVTMESSLVA P UniProtKB: Transmembrane ATP-binding protein ABC transporter cydD |
-Macromolecule #2: Component linked with the assembly of cytochrome' ABC transporter...
| Macromolecule | Name: Component linked with the assembly of cytochrome' ABC transporter ATP-binding protein CydC type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) |
| Molecular weight | Theoretical: 54.821535 KDa |
| Recombinant expression | Organism:  Mycolicibacterium smegmatis (bacteria) Mycolicibacterium smegmatis (bacteria) |
| Sequence | String: VPLLRHDPLL RLTLELLRPR LGRFLLAAAL GVLSLGSALA LAGISAWLIT RAWQMPPVLD LTVAVVAVRA LGISRGVLGY CQRLASHDS ALRAAANART GLYRKLADAP PDEAMRLPSG ELVARLGPAV DELADVLVRA LLPIVVAVVL GCAAVGVIAV I SPASAAVL ...String: VPLLRHDPLL RLTLELLRPR LGRFLLAAAL GVLSLGSALA LAGISAWLIT RAWQMPPVLD LTVAVVAVRA LGISRGVLGY CQRLASHDS ALRAAANART GLYRKLADAP PDEAMRLPSG ELVARLGPAV DELADVLVRA LLPIVVAVVL GCAAVGVIAV I SPASAAVL AVCLVVAGVV APALAARAAH ASETVAAEHR SQRDTAGMLA LEHAPELRVS GRLDSVIATF ERHHRAWGEA AD RAAAPAA VAAAMPTAAM GVSVVGAVIA GIALAPTVAP TTAAILMLLP LSAFEATTAL PDAAAQLMRS RVAARRLLEL TTP TPLRSR PDVATVDLAP GDRLAVVGPS GSGKTTMLMA IADRLNGAGG ETPQRAAVFA EDAHLFDTTV RDNLLVVRGD ATDT ELVAA LDRVGLGEWL AGLPDGLSTV LVGGAAAVSA GQRRRLLIAR ALISAFPVVL LDEPTENLDA GDARQMLEGL LTPGA LFAA DRTVVVATHH LPPGFDCPIV RCTGRLAVAG RYLGGIKAFD YKDDDDK UniProtKB: Component linked with the assembly of cytochrome' ABC transporter ATP-binding protein CydC |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.5 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 107090 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller








 Z
Z Y
Y X
X