+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Eaf3 CHD domain bound to the nucleosome | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | HDAC / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationRpd3S complex / NuA4 histone acetyltransferase complex / structural constituent of chromatin / nucleosome / nucleosome assembly / chromatin remodeling / protein heterodimerization activity / DNA repair / regulation of DNA-templated transcription / DNA binding / nucleus Similarity search - Function | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.58 Å | |||||||||
 Authors Authors | Zhang Y / Gang C | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Cell Res / Year: 2023 Journal: Cell Res / Year: 2023Title: Structural basis for nucleosome binding and catalysis by the yeast Rpd3S/HDAC holoenzyme. Authors: Yueyue Zhang / Mengxue Xu / Po Wang / Jiahui Zhou / Guangxian Wang / Shuailong Han / Gang Cai / Xuejuan Wang /  | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35449.map.gz emd_35449.map.gz | 136.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35449-v30.xml emd-35449-v30.xml emd-35449.xml emd-35449.xml | 20.6 KB 20.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_35449.png emd_35449.png | 48.3 KB | ||
| Filedesc metadata |  emd-35449.cif.gz emd-35449.cif.gz | 6.4 KB | ||
| Others |  emd_35449_half_map_1.map.gz emd_35449_half_map_1.map.gz emd_35449_half_map_2.map.gz emd_35449_half_map_2.map.gz | 134.1 MB 134.1 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35449 http://ftp.pdbj.org/pub/emdb/structures/EMD-35449 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35449 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35449 | HTTPS FTP |
-Related structure data
| Related structure data |  8ihmMC  8ihnC  8ihtC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_35449.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35449.map.gz / Format: CCP4 / Size: 144.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||
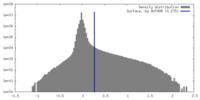
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_35449_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
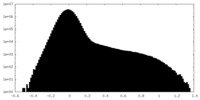
| Projections & Slices |
| ||||||||||||
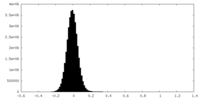
| Density Histograms |
-Half map: #1
| File | emd_35449_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
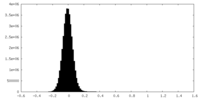
| Density Histograms |
- Sample components
Sample components
-Entire : Eaf3 CHD domain bound to the nucleosome
| Entire | Name: Eaf3 CHD domain bound to the nucleosome |
|---|---|
| Components |
|
-Supramolecule #1: Eaf3 CHD domain bound to the nucleosome
| Supramolecule | Name: Eaf3 CHD domain bound to the nucleosome / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#8 |
|---|---|
| Source (natural) | Organism: |
-Macromolecule #1: Histone H3
| Macromolecule | Name: Histone H3 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 15.331982 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ARTKQTARKS TGGKAPRKQL ATKAARKSAP ATGGV(ML3)KPHR YRPGTVALRE IRRYQKSTEL LIRKLPFQRL VREIAQ DFK TDLRFQSSAV MALQEASEAY LVALFEDTNL AAIHAKRVTI MPKDIQLARR IRGERA UniProtKB: Histone H3 |
-Macromolecule #2: Histone H4
| Macromolecule | Name: Histone H4 / type: protein_or_peptide / ID: 2 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 11.263231 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SGRGKGGKGL GKGGAKRHRK VLRDNIQGIT KPAIRRLARR GGVKRISGLI YEETRGVLKV FLENVIRDAV TYTEHAKRKT VTAMDVVYA LKRQGRTLYG FGG UniProtKB: Histone H4 |
-Macromolecule #3: Histone H2A
| Macromolecule | Name: Histone H2A / type: protein_or_peptide / ID: 3 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 13.978241 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: SGRGKQGGKT RAKAKTRSSR AGLQFPVGRV HRLLRKGNYA ERVGAGAPVY LAAVLEYLTA EILELAGNAA RDNKKTRIIP RHLQLAVRN DEELNKLLGR VTIAQGGVLP NIQSVLLPKK TESSKSAKSK UniProtKB: Histone H2A |
-Macromolecule #4: Histone H2B
| Macromolecule | Name: Histone H2B / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 13.524752 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: AKSAPAPKKG SKKAVTKTQK KDGKKRRKTR KESYAIYVYK VLKQVHPDTG ISSKAMSIMN SFVNDVFERI AGEASRLAHY NKRSTITSR EIQTAVRLLL PGELAKHAVS EGTKAVTKYT SAK UniProtKB: Histone H2B |
-Macromolecule #7: Rco1-ABR
| Macromolecule | Name: Rco1-ABR / type: protein_or_peptide / ID: 7 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 2.206506 KDa |
| Sequence | String: RRKIITSEGI ERSFKNEH |
-Macromolecule #8: Chromatin modification-related protein EAF3
| Macromolecule | Name: Chromatin modification-related protein EAF3 / type: protein_or_peptide / ID: 8 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 13.858936 KDa |
| Sequence | String: RCLAFHGPLM YEAKILKIWD PSSKMYTSIP NDKPGGSSQA TKEIKPQKLG EDESIPEEII NGKCFFIHYQ GWKSSWDEWV GYDRIRAYN EENIAMKKRL ANEAKEAKKS LLEQQKKKKL UniProtKB: Chromatin modification-related protein EAF3 |
-Macromolecule #5: DNA (164-MER)
| Macromolecule | Name: DNA (164-MER) / type: dna / ID: 5 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 50.291 KDa |
| Sequence | String: (DT)(DC)(DG)(DC)(DC)(DC)(DT)(DT)(DA)(DC) (DT)(DG)(DG)(DC)(DC)(DG)(DC)(DC)(DC)(DT) (DG)(DG)(DA)(DG)(DA)(DA)(DT)(DC)(DC) (DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA)(DG) (DG) (DC)(DC)(DG)(DC)(DT)(DC) ...String: (DT)(DC)(DG)(DC)(DC)(DC)(DT)(DT)(DA)(DC) (DT)(DG)(DG)(DC)(DC)(DG)(DC)(DC)(DC)(DT) (DG)(DG)(DA)(DG)(DA)(DA)(DT)(DC)(DC) (DC)(DG)(DG)(DT)(DG)(DC)(DC)(DG)(DA)(DG) (DG) (DC)(DC)(DG)(DC)(DT)(DC)(DA)(DA) (DT)(DT)(DG)(DG)(DT)(DC)(DG)(DT)(DA)(DG) (DA)(DC) (DA)(DG)(DC)(DT)(DC)(DT)(DA) (DG)(DC)(DA)(DC)(DC)(DG)(DC)(DT)(DT)(DA) (DA)(DA)(DC) (DG)(DC)(DA)(DC)(DG)(DT) (DA)(DC)(DG)(DC)(DG)(DC)(DT)(DG)(DT)(DC) (DC)(DC)(DC)(DC) (DG)(DC)(DG)(DT)(DT) (DT)(DT)(DA)(DA)(DC)(DC)(DG)(DC)(DC)(DA) (DA)(DG)(DG)(DG)(DG) (DA)(DT)(DT)(DA) (DC)(DT)(DC)(DC)(DC)(DT)(DA)(DG)(DT)(DC) (DT)(DC)(DC)(DA)(DG)(DG) (DC)(DA)(DC) (DG)(DT)(DG)(DT)(DC)(DA)(DG)(DA)(DT)(DA) (DT)(DA)(DT)(DA)(DC)(DA)(DT) (DC)(DC) (DT)(DG) |
-Macromolecule #6: DNA (165-MER)
| Macromolecule | Name: DNA (165-MER) / type: dna / ID: 6 / Number of copies: 1 / Classification: DNA |
|---|---|
| Source (natural) | Organism: |
| Molecular weight | Theoretical: 51.256629 KDa |
| Sequence | String: (DC)(DA)(DG)(DG)(DA)(DT)(DG)(DT)(DA)(DT) (DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC)(DA) (DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG)(DG) (DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG)(DA) (DG) (DT)(DA)(DA)(DT)(DC)(DC) ...String: (DC)(DA)(DG)(DG)(DA)(DT)(DG)(DT)(DA)(DT) (DA)(DT)(DA)(DT)(DC)(DT)(DG)(DA)(DC)(DA) (DC)(DG)(DT)(DG)(DC)(DC)(DT)(DG)(DG) (DA)(DG)(DA)(DC)(DT)(DA)(DG)(DG)(DG)(DA) (DG) (DT)(DA)(DA)(DT)(DC)(DC)(DC)(DC) (DT)(DT)(DG)(DG)(DC)(DG)(DG)(DT)(DT)(DA) (DA)(DA) (DA)(DC)(DG)(DC)(DG)(DG)(DG) (DG)(DG)(DA)(DC)(DA)(DG)(DC)(DG)(DC)(DG) (DT)(DA)(DC) (DG)(DT)(DG)(DC)(DG)(DT) (DT)(DT)(DA)(DA)(DG)(DC)(DG)(DG)(DT)(DG) (DC)(DT)(DA)(DG) (DA)(DG)(DC)(DT)(DG) (DT)(DC)(DT)(DA)(DC)(DG)(DA)(DC)(DC)(DA) (DA)(DT)(DT)(DG)(DA) (DG)(DC)(DG)(DG) (DC)(DC)(DT)(DC)(DG)(DG)(DC)(DA)(DC)(DC) (DG)(DG)(DG)(DA)(DT)(DT) (DC)(DT)(DC) (DC)(DA)(DG)(DG)(DG)(DC)(DG)(DG)(DC)(DC) (DA)(DG)(DT)(DA)(DA)(DG)(DG) (DG)(DC) (DG)(DA)(DC) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.6 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 53.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.8 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.58 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 107252 |
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller











 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)