+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3544 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM reconstruction of AaLS-13 | |||||||||
 Map data Map data | cryo-EM map of AaLS-13 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | cryo-EM / porous protein cage / icosahedral symmetry / engineered lumazine synthase / TRANSFERASE | |||||||||
| Function / homology |  Function and homology information Function and homology information6,7-dimethyl-8-ribityllumazine synthase / 6,7-dimethyl-8-ribityllumazine synthase activity / riboflavin synthase complex / riboflavin biosynthetic process / cytosol Similarity search - Function | |||||||||
| Biological species |   Aquifex aeolicus (bacteria) Aquifex aeolicus (bacteria) | |||||||||
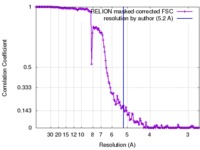
| Method | single particle reconstruction / cryo EM / Resolution: 5.2 Å | |||||||||
 Authors Authors | Sasaki E / Boehringer D | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2017 Journal: Nat Commun / Year: 2017Title: Structure and assembly of scalable porous protein cages. Authors: Eita Sasaki / Daniel Böhringer / Michiel van de Waterbeemd / Marc Leibundgut / Reinhard Zschoche / Albert J R Heck / Nenad Ban / Donald Hilvert /   Abstract: Proteins that self-assemble into regular shell-like polyhedra are useful, both in nature and in the laboratory, as molecular containers. Here we describe cryo-electron microscopy (EM) structures of ...Proteins that self-assemble into regular shell-like polyhedra are useful, both in nature and in the laboratory, as molecular containers. Here we describe cryo-electron microscopy (EM) structures of two versatile encapsulation systems that exploit engineered electrostatic interactions for cargo loading. We show that increasing the number of negative charges on the lumenal surface of lumazine synthase, a protein that naturally assembles into a ∼1-MDa dodecahedron composed of 12 pentamers, induces stepwise expansion of the native protein shell, giving rise to thermostable ∼3-MDa and ∼6-MDa assemblies containing 180 and 360 subunits, respectively. Remarkably, these expanded particles assume unprecedented tetrahedrally and icosahedrally symmetric structures constructed entirely from pentameric units. Large keyhole-shaped pores in the shell, not present in the wild-type capsid, enable diffusion-limited encapsulation of complementarily charged guests. The structures of these supercharged assemblies demonstrate how programmed electrostatic effects can be effectively harnessed to tailor the architecture and properties of protein cages. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3544.map.gz emd_3544.map.gz | 36.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3544-v30.xml emd-3544-v30.xml emd-3544.xml emd-3544.xml | 10 KB 10 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_3544_fsc.xml emd_3544_fsc.xml | 14.5 KB | Display |  FSC data file FSC data file |
| Images |  emd_3544.png emd_3544.png | 107.6 KB | ||
| Filedesc metadata |  emd-3544.cif.gz emd-3544.cif.gz | 4.9 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3544 http://ftp.pdbj.org/pub/emdb/structures/EMD-3544 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3544 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3544 | HTTPS FTP |
-Validation report
| Summary document |  emd_3544_validation.pdf.gz emd_3544_validation.pdf.gz | 283.4 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3544_full_validation.pdf.gz emd_3544_full_validation.pdf.gz | 282.5 KB | Display | |
| Data in XML |  emd_3544_validation.xml.gz emd_3544_validation.xml.gz | 14.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3544 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3544 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3544 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3544 | HTTPS FTP |
-Related structure data
| Related structure data |  5mq7MC  3538C  3543C  5mppC  5mq3C M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3544.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3544.map.gz / Format: CCP4 / Size: 282.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | cryo-EM map of AaLS-13 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.39 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : AaLS-13
| Entire | Name: AaLS-13 |
|---|---|
| Components |
|
-Supramolecule #1: AaLS-13
| Supramolecule | Name: AaLS-13 / type: complex / ID: 1 / Parent: 0 / Macromolecule list: all |
|---|---|
| Source (natural) | Organism:   Aquifex aeolicus (bacteria) Aquifex aeolicus (bacteria) |
-Macromolecule #1: 6,7-dimethyl-8-ribityllumazine synthase
| Macromolecule | Name: 6,7-dimethyl-8-ribityllumazine synthase / type: protein_or_peptide / ID: 1 / Number of copies: 360 / Enantiomer: LEVO / EC number: 6,7-dimethyl-8-ribityllumazine synthase |
|---|---|
| Source (natural) | Organism:   Aquifex aeolicus (bacteria) / Strain: VF5 Aquifex aeolicus (bacteria) / Strain: VF5 |
| Molecular weight | Theoretical: 17.569855 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEIYEGKLTA EGLRFGIVAS RFNHALVGRL VEGAIDCIVR HGGREEDITL VCVPGSWEIP VAAGELARKE DIDAVIAIGV LIEGAEPHF DYIASEVSKG LANLSLELRK PISFGDITDD ELEEAIECAG TEHGNKGWEA ALSAIEMANL FKSLRLEHHH H H UniProtKB: 6,7-dimethyl-8-ribityllumazine synthase |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: FEI FALCON II (4k x 4k) / Average electron dose: 4.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller