[English] 日本語
 Yorodumi
Yorodumi- EMDB-35416: Cryo-EM structure of the Clr6S (Clr6-HDAC) complex from S. pombe -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-EM structure of the Clr6S (Clr6-HDAC) complex from S. pombe | |||||||||
 Map data Map data | Cryo-EM map of Clr6-HDAC complex | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Rpd3S / Histone deacetylase / HDAC / Clr6 / Rpd3 / Clr6S / Clr6 HDAC / TRANSCRIPTION | |||||||||
| Function / homology |  Function and homology information Function and homology informationhistone H4K16 deacetylase activity, hydrolytic mechanism / histone H4K5 deacetylase activity, hydrolytic mechanism / histone H4K8 deacetylase activity, hydrolytic mechanism / histone H3K9 deacetylase activity, hydrolytic mechanism / histone H3K14 deacetylase activity, hydrolytic mechanism / HDACs deacetylate histones / histone H4K12 deacetylase activity, hydrolytic mechanism / SUMOylation of chromatin organization proteins / H3-H4 histone complex chaperone activity / Rpd3L complex ...histone H4K16 deacetylase activity, hydrolytic mechanism / histone H4K5 deacetylase activity, hydrolytic mechanism / histone H4K8 deacetylase activity, hydrolytic mechanism / histone H3K9 deacetylase activity, hydrolytic mechanism / histone H3K14 deacetylase activity, hydrolytic mechanism / HDACs deacetylate histones / histone H4K12 deacetylase activity, hydrolytic mechanism / SUMOylation of chromatin organization proteins / H3-H4 histone complex chaperone activity / Rpd3L complex / Rpd3L-Expanded complex / Rpd3S complex / histone deacetylase / protein lysine deacetylase activity / histone deacetylase activity / DNA repair-dependent chromatin remodeling / NuA4 histone acetyltransferase complex / Sin3-type complex / epigenetic regulation of gene expression / transcription initiation-coupled chromatin remodeling / heterochromatin formation / mitotic spindle / transcription corepressor activity / histone binding / molecular adaptor activity / chromatin remodeling / DNA repair / regulation of DNA-templated transcription / chromatin / regulation of transcription by RNA polymerase II / negative regulation of transcription by RNA polymerase II / nucleus / metal ion binding / cytosol / cytoplasm Similarity search - Function | |||||||||
| Biological species |   | |||||||||
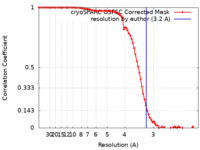
| Method | single particle reconstruction / cryo EM / Resolution: 3.2 Å | |||||||||
 Authors Authors | Zhang HQ / Wang X / Wang YN / Liu SM / Zhang Y / Xu K / Ji LT / Kornberg RD | |||||||||
| Funding support | 1 items
| |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2023 Journal: Proc Natl Acad Sci U S A / Year: 2023Title: Class I histone deacetylase complex: Structure and functional correlates. Authors: Xiao Wang / Yannan Wang / Simiao Liu / Yi Zhang / Ke Xu / Liting Ji / Roger D Kornberg / Heqiao Zhang /   Abstract: The Clr6S complex, a class I histone deacetylase complex, functions as a zinc-dependent enzyme to remove acetyl groups from lysine residues in histone tails. We report here the cryo-EM structure of ...The Clr6S complex, a class I histone deacetylase complex, functions as a zinc-dependent enzyme to remove acetyl groups from lysine residues in histone tails. We report here the cryo-EM structure of Clr6S alone and a cryo-EM map of Clr6S in complex with a nucleosome. The active center, revealed at near-atomic resolution, includes features important for catalysis-A water molecule coordinated by zinc, the likely nucleophile for attack on the acetyl-lysine bond, and a loop that may position the substrate for catalysis. The cryo-EM map in the presence of a nucleosome reveals multiple Clr6S-nucleosome contacts and a high degree of relative motion of Clr6S and the nucleosome. Such flexibility may be attributed to interaction at a site in the flexible histone tail and is likely important for the function of the deacetylase, which acts at multiple sites in other histone tails. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_35416.map.gz emd_35416.map.gz | 56.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-35416-v30.xml emd-35416-v30.xml emd-35416.xml emd-35416.xml | 23.2 KB 23.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_35416_fsc.xml emd_35416_fsc.xml | 9.3 KB | Display |  FSC data file FSC data file |
| Images |  emd_35416.png emd_35416.png | 29.3 KB | ||
| Filedesc metadata |  emd-35416.cif.gz emd-35416.cif.gz | 7.9 KB | ||
| Others |  emd_35416_half_map_1.map.gz emd_35416_half_map_1.map.gz emd_35416_half_map_2.map.gz emd_35416_half_map_2.map.gz | 55.4 MB 55.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-35416 http://ftp.pdbj.org/pub/emdb/structures/EMD-35416 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35416 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-35416 | HTTPS FTP |
-Validation report
| Summary document |  emd_35416_validation.pdf.gz emd_35416_validation.pdf.gz | 1018.6 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_35416_full_validation.pdf.gz emd_35416_full_validation.pdf.gz | 1018.2 KB | Display | |
| Data in XML |  emd_35416_validation.xml.gz emd_35416_validation.xml.gz | 16 KB | Display | |
| Data in CIF |  emd_35416_validation.cif.gz emd_35416_validation.cif.gz | 20.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35416 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35416 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35416 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-35416 | HTTPS FTP |
-Related structure data
| Related structure data |  8ifgMC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_35416.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_35416.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Cryo-EM map of Clr6-HDAC complex | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.06 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map
| File | emd_35416_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: half map
| File | emd_35416_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Cryo-EM structure of the Clr6-HDAC (Clr6S) complex from S. pombe
| Entire | Name: Cryo-EM structure of the Clr6-HDAC (Clr6S) complex from S. pombe |
|---|---|
| Components |
|
-Supramolecule #1: Cryo-EM structure of the Clr6-HDAC (Clr6S) complex from S. pombe
| Supramolecule | Name: Cryo-EM structure of the Clr6-HDAC (Clr6S) complex from S. pombe type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#6 |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 600 KDa |
-Macromolecule #1: RbAp48-related WD40 repeat-containing protein prw1
| Macromolecule | Name: RbAp48-related WD40 repeat-containing protein prw1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 48.528926 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAVSAVPHPS KQAQASEEGI NQEKCINEEY KIWKKNSPFL YDLIITRALE WPCMSLQWYP EQQIFAEHGY TEQKMFLGVR ADVGKYLLA VASIQLPYLN QTVPPTTMEG ASAGDESSLR VNISNLYSHP ESVCSAKLMP QDDSCVATVG NYHNDVLVFD K ESFESYSS ...String: MAVSAVPHPS KQAQASEEGI NQEKCINEEY KIWKKNSPFL YDLIITRALE WPCMSLQWYP EQQIFAEHGY TEQKMFLGVR ADVGKYLLA VASIQLPYLN QTVPPTTMEG ASAGDESSLR VNISNLYSHP ESVCSAKLMP QDDSCVATVG NYHNDVLVFD K ESFESYSS ASESPLKPKY RLTKHTQPCT SVCWNFLSKG TLVSGSQDAT LSCWDLNAYN ESDSASVLKV HISSHEKQVS DV RFHYKHQ DLLASVSYDQ YLHVHDIRRP DASTKPARSV HAHSGPIHSV AFNPHNDFIL ATCSTDKTIA LWDLRNLNQR LHT LEGHED IVTKISFSPH EEPILASTSA DRRTLVWDLS RIGEDQPAEE AQDGPPELLF MHGGHTSCTI DMDWCPNYNW TMAT AAEDN ILQIWTPSRS IWGNEQLEED ATAYLS UniProtKB: RbAp48-related WD40 repeat-containing protein prw1 |
-Macromolecule #2: Paired amphipathic helix protein pst2
| Macromolecule | Name: Paired amphipathic helix protein pst2 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 125.011609 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MEQTLAILKN DNSTLVAEMQ NQLVHDFSPN GTALPELDIK AFVQKLGQRL CHRPYVYSAF MDVVKALHNE IVDFPGFIER ISVILRDYP DLLEYLNIFL PSSYKYLLSN SGANFTLQFT TPSGPVSTPS TYVATYNDLP CTYHRAIGFV SRVRRALLSN P EQFFKLQD ...String: MEQTLAILKN DNSTLVAEMQ NQLVHDFSPN GTALPELDIK AFVQKLGQRL CHRPYVYSAF MDVVKALHNE IVDFPGFIER ISVILRDYP DLLEYLNIFL PSSYKYLLSN SGANFTLQFT TPSGPVSTPS TYVATYNDLP CTYHRAIGFV SRVRRALLSN P EQFFKLQD SLRKFKNSEC SLSELQTIVT SLLAEHPSLA HEFHNFLPSS IFFGSKPPLG SFPLRGIQSS QFTLSNISDL LS QSRPDNL SPFSHLSNES SDFFKNVKNV LTDVETYHEF LKLLNLYVQG IIDRNILVSR GFGFLKSNSG LWRSFLSLTS LSP EEFLSV YNSACSDFPE CGPSYRLLPV EERNISCSGR DDFAWGILND DWVSHPTWAS EESGFIVQRK TPYEEAMTKL EEER YEFDR HIEATSWTIK SLKKIQNRIN ELPEEERETY TLEEGLGLPS KSIYKKTIKL VYTSEHAEEM FKALERMPCL TLPLV ISRL EEKNEEWKSV KRSLQPGWRS IEFKNYDKSL DSQCVYFKAR DKKNVSSKFL LAEADILRSQ AKLHFPLRSR SAFEFS FVY DNEIVLFDTC YMVCTYIVCN SPSGLKKVEH FFKNILPLHF GLEKDKFSIF LDQVFRGPDY DVNAPNIVGN KPVRRKR SN SITQLTEFVK QPKINGQRES RSAAAARKKE ESGNKSQSNS QNSLSDESGN VTPVSKKQLS QPAAAIKASL KYPSHPDS L LEHQDHAGDT ENEMHDDVDK EQFGYSSMYV FFRLFNLLYE RLYELQRLED QVSIIQQRII PNPVSQKQKI WRDRWNDLS DVPDEKTHYE NTYVMILRLI YGIVDQSAFE DYLRFYYGNK AYKIYTIDKL VWSAAKQVHH IVSDGKYKFV TSLVEQNSSA SPKKNYDDF LYRLEIEKLL NPDEILFRFC WINKFKSFGI KIMKRANLIV DQSLDTQRRV WKKYVQNYRI QKLTEEISYK N YRCPFLCR NIEKERTVEQ LVSRLQTKLL RSAELVSGLQ AKLCLDSFKL LYLPRTEDSY IDASYLRLRD TDFLDCQNKR KQ RWRNRWE SLLKSVRGTS DNTAEVNFDA DINALFIP UniProtKB: Paired amphipathic helix protein pst2 |
-Macromolecule #3: Histone deacetylase clr6
| Macromolecule | Name: Histone deacetylase clr6 / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO / EC number: histone deacetylase |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.773371 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: KKKVSYFYDE DVGNYHYGPQ HPMKPHRVRM VHNLVVNYNL YEKLNVITPV RATRNDMTRC HTDEYIEFLW RVTPDTMEKF QPHQLKFNV GDDCPVFDGL YEFCSISAGG SIGAAQELNS GNAEIAINWA GGLHHAKKRE ASGFCYVNDI ALAALELLKY H QRVLYIDI ...String: KKKVSYFYDE DVGNYHYGPQ HPMKPHRVRM VHNLVVNYNL YEKLNVITPV RATRNDMTRC HTDEYIEFLW RVTPDTMEKF QPHQLKFNV GDDCPVFDGL YEFCSISAGG SIGAAQELNS GNAEIAINWA GGLHHAKKRE ASGFCYVNDI ALAALELLKY H QRVLYIDI DVHHGDGVEE FFYTTDRVMT CSFHKFGEYF PGTGHIKDTG IGTGKNYAVN VPLRDGIDDE SYESVFKPVI SH IMQWFRP EAVILQCGTD SLAGDRLGCF NLSMKGHSMC VDFVKSFNLP MICVGGGGYT VRNVARVWTY ETGLLAGEEL DEN LPYNDY LQYYGPDYKL NVLSNNMENH NTRQYLDSIT SEIIENLRNL SFAPSVQMHK TPGDFTFENA EKQNIAKEEI MDER V UniProtKB: Histone deacetylase clr6 |
-Macromolecule #4: Chromatin modification-related protein eaf3
| Macromolecule | Name: Chromatin modification-related protein eaf3 / type: protein_or_peptide / ID: 4 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 39.191199 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MAVSYKVNER VLCFHGPLLY EAKIVDTEMK GDVTTYLIHY KGWKNSWDEW VEQDRILQWT EENLKTQKEL KNAAISTRQK PTSKKSASS TSKHDSTGVK TSGKRSRESS TVTVDGDSHE LPSRIKTQKS ESPIPQQVKR DGTTDAKNEE TTKPENNEKD D FEEEPPLP ...String: MAVSYKVNER VLCFHGPLLY EAKIVDTEMK GDVTTYLIHY KGWKNSWDEW VEQDRILQWT EENLKTQKEL KNAAISTRQK PTSKKSASS TSKHDSTGVK TSGKRSRESS TVTVDGDSHE LPSRIKTQKS ESPIPQQVKR DGTTDAKNEE TTKPENNEKD D FEEEPPLP KHKISVPDVL KLWLVDDWEN ITKNQQLIAI PRNPTVRAAI AAFRESKISH LNNEIDVDVF EQAMAGLVIY FN KCLGNML LYRFERQQYL EIRQQYPDTE MCDLYGVEHL IRLFVSLPEL IDRTNMDSQS IECLLNYIEE FLKYLVLHKD EYF IKEYQN APPNYRSLVG V UniProtKB: Chromatin modification-related protein eaf3 |
-Macromolecule #5: Cph1
| Macromolecule | Name: Cph1 / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 45.046082 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MASSINNSSQ PTVPSISNNS HGDSFVNEGP PSNFKNNSLT SSTHSSTDHV NVLPISQDKE MDISSPVKKQ KASYSNKSPN KAPIQKSRG SSLKSHLETE SQQTPVKRRR RKATIRNVDY CSACGGRGLF ICCEGCPCSF HLSCLEPPLT PENIPEGSWF C VTCSIKSH ...String: MASSINNSSQ PTVPSISNNS HGDSFVNEGP PSNFKNNSLT SSTHSSTDHV NVLPISQDKE MDISSPVKKQ KASYSNKSPN KAPIQKSRG SSLKSHLETE SQQTPVKRRR RKATIRNVDY CSACGGRGLF ICCEGCPCSF HLSCLEPPLT PENIPEGSWF C VTCSIKSH HPPKHPLSIW SQLYDWIDSQ NPSQYRLPDD LVHYFHGISR GDTGAYKETE GEMDTDEFSA LPTGSSITNL AY CGYCSKP SMGACWVYGC QLCDTFYHKN CKEHAKKCSH DSIGKKGMRV PKNAVVIRTP LVLDTTSNTL NPKVMISGWQ FLM GEFPSD ELLYFPRLPV SCLYKVSEDG LIKDFLYAIG IEAKKFNNER KKRELEVIPP DVKSALLPAR THPNLPIALR TLFN KART UniProtKB: Uncharacterized protein C16C9.05 |
-Macromolecule #6: Cph2
| Macromolecule | Name: Cph2 / type: protein_or_peptide / ID: 6 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 68.871703 KDa |
| Recombinant expression | Organism:  Trichoplusia ni (cabbage looper) Trichoplusia ni (cabbage looper) |
| Sequence | String: MDAKPWNHTS EAFQASILED LKIIQKAGAE RNAKSSHGSI NSRSASPNKA TSRRNRAQNG NSNGRASVDN SDDGSKDDLD YSPSVKRKH VNGEGAEKGD HDTSNNGPSI TKLRRKVRRT YDTKDGFVAW NTLDDDFRPI VPDQERSRKI NPQKGNNNNL L KENKSLKT ...String: MDAKPWNHTS EAFQASILED LKIIQKAGAE RNAKSSHGSI NSRSASPNKA TSRRNRAQNG NSNGRASVDN SDDGSKDDLD YSPSVKRKH VNGEGAEKGD HDTSNNGPSI TKLRRKVRRT YDTKDGFVAW NTLDDDFRPI VPDQERSRKI NPQKGNNNNL L KENKSLKT TAKDLSDISS SSMKKANNSS KPLFSGKLTF KANIPVPTSE VVTENNVTRN VTVYSNQKHL GNESENFNDM EG RAEDISS NELLPTPEEY PYRYNNDYCS ACHGPGNFLC CETCPNSFHF TCIDPPIEEK NLPDDAWYCN ECKHHSLYNE LDE QEELES NVKEEGTMVD VWMQLCTYID SHNPIQFHLP HSISSFFRGV GSGVMGEYIE TDVLKHLKSS RRSNGEERDP LLLK SKSGT PILCFRCHKS ALVSQSILAC DYCNSYWHPD CLNPPLATLP SNLRKWKCPN HSDHVTPRYR LPEKAKVIRV GLPRG FKNK GNIVIDENED EPSVQTIQLQ GKIRVVPSKP FKLNFLEQIR DNVINLRKMV EQDEQLCIET FSKFDFYATR DCELPL RIL CDVANDNLEN DDYVLALRDL LRISKWDPNQ PVPAPFDLAN LLSY UniProtKB: Uncharacterized protein C2F7.07c |
-Macromolecule #7: ZINC ION
| Macromolecule | Name: ZINC ION / type: ligand / ID: 7 / Number of copies: 3 / Formula: ZN |
|---|---|
| Molecular weight | Theoretical: 65.409 Da |
-Macromolecule #8: water
| Macromolecule | Name: water / type: ligand / ID: 8 / Number of copies: 1 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller





 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)