+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Structure of tetradecameric RdrA ring | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Cryoelectron microscopy / adenosine triphosphatase / IMMUNE SYSTEM | |||||||||
| Function / homology | P-loop containing nucleoside triphosphate hydrolase / Archaeal ATPase Function and homology information Function and homology information | |||||||||
| Biological species |  | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.03 Å | |||||||||
 Authors Authors | Gao Y | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2023 Journal: Cell / Year: 2023Title: Molecular basis of RADAR anti-phage supramolecular assemblies. Authors: Yina Gao / Xiu Luo / Peipei Li / Zhaolong Li / Feng Ye / Songqing Liu / Pu Gao /  Abstract: Adenosine-to-inosine RNA editing has been proposed to be involved in a bacterial anti-phage defense system called RADAR. RADAR contains an adenosine triphosphatase (RdrA) and an adenosine deaminase ...Adenosine-to-inosine RNA editing has been proposed to be involved in a bacterial anti-phage defense system called RADAR. RADAR contains an adenosine triphosphatase (RdrA) and an adenosine deaminase (RdrB). Here, we report cryo-EM structures of RdrA, RdrB, and currently identified RdrA-RdrB complexes in the presence or absence of RNA and ATP. RdrB assembles into a dodecameric cage with catalytic pockets facing outward, while RdrA adopts both autoinhibited tetradecameric and activation-competent heptameric rings. Structural and functional data suggest a model in which RNA is loaded through the bottom section of the RdrA ring and translocated along its inner channel, a process likely coupled with ATP-binding status. Intriguingly, up to twelve RdrA rings can dock one RdrB cage with precise alignments between deaminase catalytic pockets and RNA-translocation channels, indicative of enzymatic coupling of RNA translocation and deamination. Our data uncover an interesting mechanism of enzymatic coupling and anti-phage defense through supramolecular assemblies. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34964.map.gz emd_34964.map.gz | 302.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34964-v30.xml emd-34964-v30.xml emd-34964.xml emd-34964.xml | 13.9 KB 13.9 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34964.png emd_34964.png | 119.5 KB | ||
| Others |  emd_34964_half_map_1.map.gz emd_34964_half_map_1.map.gz emd_34964_half_map_2.map.gz emd_34964_half_map_2.map.gz | 256.5 MB 257.9 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34964 http://ftp.pdbj.org/pub/emdb/structures/EMD-34964 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34964 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34964 | HTTPS FTP |
-Validation report
| Summary document |  emd_34964_validation.pdf.gz emd_34964_validation.pdf.gz | 969.3 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34964_full_validation.pdf.gz emd_34964_full_validation.pdf.gz | 968.8 KB | Display | |
| Data in XML |  emd_34964_validation.xml.gz emd_34964_validation.xml.gz | 16.9 KB | Display | |
| Data in CIF |  emd_34964_validation.cif.gz emd_34964_validation.cif.gz | 20.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34964 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34964 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34964 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34964 | HTTPS FTP |
-Related structure data
| Related structure data |  8hr9MC  8hr7C  8hr8C  8hraC  8hrbC  8hrcC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34964.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34964.map.gz / Format: CCP4 / Size: 325 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.04 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34964_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
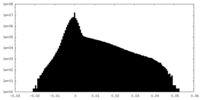
| Projections & Slices |
| ||||||||||||
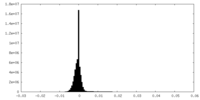
| Density Histograms |
-Half map: #1
| File | emd_34964_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
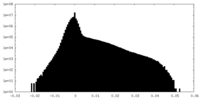
| Projections & Slices |
| ||||||||||||
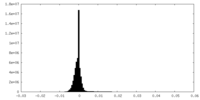
| Density Histograms |
- Sample components
Sample components
-Entire : RdrA
| Entire | Name: RdrA |
|---|---|
| Components |
|
-Supramolecule #1: RdrA
| Supramolecule | Name: RdrA / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  |
-Macromolecule #1: Archaeal ATPase
| Macromolecule | Name: Archaeal ATPase / type: protein_or_peptide / ID: 1 / Number of copies: 14 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 107.132523 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTDSVQTETT EGKIIINLFA PNLPGSTKED DLIQKSLRDQ LVESIRNSIA YPDTDKFAGL TRFIDESGRN VFFVDGTRGA GKTTFINSV VKSLNSDQDD VKVNIKCLPT IDPTKLPRHE PILVTVTARL NKMVSDKLKG YWASNDYRKQ KEQWQNHLAQ L QRGLHLLT ...String: MTDSVQTETT EGKIIINLFA PNLPGSTKED DLIQKSLRDQ LVESIRNSIA YPDTDKFAGL TRFIDESGRN VFFVDGTRGA GKTTFINSV VKSLNSDQDD VKVNIKCLPT IDPTKLPRHE PILVTVTARL NKMVSDKLKG YWASNDYRKQ KEQWQNHLAQ L QRGLHLLT DKEYKPEYFS DALKLDAQLD YSIGGQDLSE IFEELVKRAC EILDCKAILI TFDDIDTQFD AGWDVLESIR KF FNSRKLV VVATGDLRLY SQLIRGKQYE NYSKTLLEQE KESVRLAERG YMVEHLEQQY LLKLFPVQKR IQLKTMLQLV GEK GKAGKE EIKVKTEPGM QDIDAIDVRQ AIGDAVREGL NLREGSDADM YVNELLKQPV RLLMQVLQDF YTKKYHATSV KLDG KQSRN ERPNELSVPN LLRNALYGSM LSSIYRAGLN YEQHRFGMDS LCKDIFTYVK QDRDFNTGFY LRPQSESEAL RNCSI YLAS QVSENCQGSL SKFLQMLLVG CGSVSIFNQF VTELARAEND REKFEQLISE YVAYMSVGRI ESASHWANRC CAVVAN SPN DEKIGVFLGM VQLNRKSRQH MPGGYKKFNI DTENGLAKAA MASSLSTVAS NNLMDFCSVF NLIGAIADIS ACRCERS AI TNAFNKVIAQ TTCIVPPWSE AAVRAEMKGS SKSADNDAAV LDVDLDPKDD GVIDESQQDD ATEFSDAITK VEQWLKNV N EIEIGIRPSA LLIGKVWSRF YFNLNNVADQ HKTRLYRNAE HGRMASQSNA AKIMRFNVLA FLHAVLVEES LYHSVSDRE YIGEGLRLNP VTSVDEFEKK IKIIGEKLKA DNKTWKNTHP LFFLLISCPI LHPFIFPVGG INCSVKALNK ETSFNKLIDE IVGDKLLSD EEWDYLTKNN DQKTNTRQQI FQNTITSLNS STIVGASYDK DTPARKTKSP LLGDSEEK UniProtKB: Archaeal ATPase |
-Macromolecule #2: ADENOSINE-5'-TRIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-TRIPHOSPHATE / type: ligand / ID: 2 / Number of copies: 10 / Formula: ATP |
|---|---|
| Molecular weight | Theoretical: 507.181 Da |
| Chemical component information |  ChemComp-ATP: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.5 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: NONE |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 3.03 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 219191 |
| Initial angle assignment | Type: PROJECTION MATCHING |
| Final angle assignment | Type: PROJECTION MATCHING |
 Movie
Movie Controller
Controller












 Z
Z Y
Y X
X