+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | C5aR1-Gi-C5a protein complex | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | GPCR / C5aR1 / C5a / complement / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationcomplement component C5a signaling pathway / presynapse organization / regulation of tau-protein kinase activity / complement component C5a receptor activity / response to peptidoglycan / Terminal pathway of complement / membrane attack complex / sensory perception of chemical stimulus / Activation of C3 and C5 / complement receptor mediated signaling pathway ...complement component C5a signaling pathway / presynapse organization / regulation of tau-protein kinase activity / complement component C5a receptor activity / response to peptidoglycan / Terminal pathway of complement / membrane attack complex / sensory perception of chemical stimulus / Activation of C3 and C5 / complement receptor mediated signaling pathway / negative regulation of macrophage chemotaxis / G-protein activation / Activation of the phototransduction cascade / Glucagon-type ligand receptors / Thromboxane signalling through TP receptor / Sensory perception of sweet, bitter, and umami (glutamate) taste / G beta:gamma signalling through PI3Kgamma / G beta:gamma signalling through CDC42 / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Ca2+ pathway / G alpha (z) signalling events / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / complement activation, alternative pathway / Adrenaline,noradrenaline inhibits insulin secretion / ADP signalling through P2Y purinoceptor 12 / positive regulation of neutrophil chemotaxis / G alpha (q) signalling events / chemokine activity / Thrombin signalling through proteinase activated receptors (PARs) / G alpha (i) signalling events / alkylglycerophosphoethanolamine phosphodiesterase activity / photoreceptor outer segment membrane / endopeptidase inhibitor activity / spectrin binding / positive regulation of macrophage chemotaxis / amyloid-beta clearance / positive regulation of vascular endothelial growth factor production / photoreceptor outer segment / T cell migration / D2 dopamine receptor binding / Adenylate cyclase inhibitory pathway / positive regulation of protein localization to cell cortex / cellular defense response / regulation of cAMP-mediated signaling / complement activation, classical pathway / G protein-coupled serotonin receptor binding / cellular response to forskolin / positive regulation of chemokine production / regulation of mitotic spindle organization / cardiac muscle cell apoptotic process / neutrophil chemotaxis / photoreceptor inner segment / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / positive regulation of epithelial cell proliferation / Peptide ligand-binding receptors / secretory granule membrane / Regulation of Complement cascade / Regulation of insulin secretion / G protein-coupled receptor binding / G protein-coupled receptor activity / astrocyte activation / microglial cell activation / mRNA transcription by RNA polymerase II / G-protein beta/gamma-subunit complex binding / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / response to peptide hormone / cognition / ADP signalling through P2Y purinoceptor 12 / Adrenaline,noradrenaline inhibits insulin secretion / G alpha (z) signalling events / ADORA2B mediated anti-inflammatory cytokines production / positive regulation of angiogenesis / sensory perception of taste / chemotaxis / GPER1 signaling / GDP binding / G-protein beta-subunit binding / heterotrimeric G-protein complex / signaling receptor complex adaptor activity / apical part of cell / GTPase binding / retina development in camera-type eye / cell cortex / phospholipase C-activating G protein-coupled receptor signaling pathway / positive regulation of cytosolic calcium ion concentration / cellular response to hypoxia / midbody / cell body / G alpha (i) signalling events / G alpha (s) signalling events / Hydrolases; Acting on acid anhydrides; Acting on GTP to facilitate cellular and subcellular movement / basolateral plasma membrane / killing of cells of another organism / cell population proliferation / Extra-nuclear estrogen signaling / positive regulation of ERK1 and ERK2 cascade / cell surface receptor signaling pathway Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.0 Å | |||||||||
 Authors Authors | Wang Y / Liu W / Xu Y / Zhuang Y / Xu HE | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Chem Biol / Year: 2023 Journal: Nat Chem Biol / Year: 2023Title: Revealing the signaling of complement receptors C3aR and C5aR1 by anaphylatoxins. Authors: Yue Wang / Weiyi Liu / Youwei Xu / Xinheng He / Qingning Yuan / Ping Luo / Wenjia Fan / Jingpeng Zhu / Xinyue Zhang / Xi Cheng / Yi Jiang / H Eric Xu / Youwen Zhuang /  Abstract: The complement receptors C3aR and C5aR1, whose signaling is selectively activated by anaphylatoxins C3a and C5a, are important regulators of both innate and adaptive immune responses. Dysregulations ...The complement receptors C3aR and C5aR1, whose signaling is selectively activated by anaphylatoxins C3a and C5a, are important regulators of both innate and adaptive immune responses. Dysregulations of C3aR and C5aR1 signaling lead to multiple inflammatory disorders, including sepsis, asthma and acute respiratory distress syndrome. The mechanism underlying endogenous anaphylatoxin recognition and activation of C3aR and C5aR1 remains elusive. Here we reported the structures of C3a-bound C3aR and C5a-bound C5aR1 as well as an apo-C3aR structure. These structures, combined with mutagenesis analysis, reveal a conserved recognition pattern of anaphylatoxins to the complement receptors that is different from chemokine receptors, unique pocket topologies of C3aR and C5aR1 that mediate ligand selectivity, and a common mechanism of receptor activation. These results provide crucial insights into the molecular understanding of C3aR and C5aR1 signaling and structural templates for rational drug design for treating inflammation disorders. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34846.map.gz emd_34846.map.gz | 51.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34846-v30.xml emd-34846-v30.xml emd-34846.xml emd-34846.xml | 19.7 KB 19.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34846.png emd_34846.png | 33.2 KB | ||
| Filedesc metadata |  emd-34846.cif.gz emd-34846.cif.gz | 6.4 KB | ||
| Others |  emd_34846_half_map_1.map.gz emd_34846_half_map_1.map.gz emd_34846_half_map_2.map.gz emd_34846_half_map_2.map.gz | 46.2 MB 46.2 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34846 http://ftp.pdbj.org/pub/emdb/structures/EMD-34846 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34846 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34846 | HTTPS FTP |
-Validation report
| Summary document |  emd_34846_validation.pdf.gz emd_34846_validation.pdf.gz | 730.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34846_full_validation.pdf.gz emd_34846_full_validation.pdf.gz | 730.4 KB | Display | |
| Data in XML |  emd_34846_validation.xml.gz emd_34846_validation.xml.gz | 11.8 KB | Display | |
| Data in CIF |  emd_34846_validation.cif.gz emd_34846_validation.cif.gz | 13.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34846 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34846 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34846 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34846 | HTTPS FTP |
-Related structure data
| Related structure data |  8hk5MC  8hk2C  8hk3C C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34846.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34846.map.gz / Format: CCP4 / Size: 59.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.824 Å | ||||||||||||||||||||||||||||||||||||
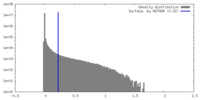
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #1
| File | emd_34846_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
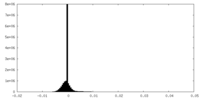
| Density Histograms |
-Half map: #2
| File | emd_34846_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : C5aR1-Gi-C5a protein complex
| Entire | Name: C5aR1-Gi-C5a protein complex |
|---|---|
| Components |
|
-Supramolecule #1: C5aR1-Gi-C5a protein complex
| Supramolecule | Name: C5aR1-Gi-C5a protein complex / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 160 KDa |
-Macromolecule #1: C5a anaphylatoxin chemotactic receptor 1
| Macromolecule | Name: C5a anaphylatoxin chemotactic receptor 1 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 39.372375 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MDSFNYTTPD YGHYDDKDTL DLNTPVDKTS NTLRVPDILA LVIFAVVFLV GVLGNALVVW VTAFEAKRTI NAIWFLNLAV ADFLSCLAL PILFTSIVQH HHWPFGGAAC SILPSLILLN MYASILLLAT ISADRFLLVF KPIWCQNFRG AGLAWIACAV A WGLALLLT ...String: MDSFNYTTPD YGHYDDKDTL DLNTPVDKTS NTLRVPDILA LVIFAVVFLV GVLGNALVVW VTAFEAKRTI NAIWFLNLAV ADFLSCLAL PILFTSIVQH HHWPFGGAAC SILPSLILLN MYASILLLAT ISADRFLLVF KPIWCQNFRG AGLAWIACAV A WGLALLLT IPSFLYRVVR EEYFPPKVLC GVDYSHDKRR ERAVAIVRLV LGFLWPLLTL TICYTFILLR TWSRRATRST KT LKVVVAV VASFFIFWLP YQVTGIMMSF LEPSSPTFLL LKKLDSLCVS FAYINCCINP IIYVVAGQGF QGRLRKSLPS LLR NVLTEE SVVRESKSFT RSTVDTMAQK TQAV UniProtKB: C5a anaphylatoxin chemotactic receptor 1 |
-Macromolecule #2: Complement C5
| Macromolecule | Name: Complement C5 / type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 8.288676 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: TLQKKIEEIA AKYKHSVVKK CCYDGACVNN DETCEQRAAR ISLGPRCIKA FTECCVVASQ LRANISHKDM QLGR UniProtKB: Complement C5 |
-Macromolecule #3: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.313863 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: GCTLSAEDKA AVERSKMIDR NLREDGEKAA REVKLLLLGA GESGKSTIVK QMKIIHEAGY SEEECKQYKA VVYSNTIQSI IAIIRAMGR LKIDFGDSAR ADDARQLFVL AGAAEEGFMT AELAGVIKRL WKDSGVQACF NRSREYQLND SAAYYLNDLD R IAQPNYIP ...String: GCTLSAEDKA AVERSKMIDR NLREDGEKAA REVKLLLLGA GESGKSTIVK QMKIIHEAGY SEEECKQYKA VVYSNTIQSI IAIIRAMGR LKIDFGDSAR ADDARQLFVL AGAAEEGFMT AELAGVIKRL WKDSGVQACF NRSREYQLND SAAYYLNDLD R IAQPNYIP TQQDVLRTRV KTTGIVETHF TFKDLHFKMF DVGAQRSERK KWIHCFEGVT AIIFCVALSD YDLVLAEDEE MN RMHESMK LFDSICNNKW FTDTSIILFL NKKDLFEEKI KKSPLTICYP EYAGSNTYEE AAAYIQCQFE DLNKRKDTKE IYT HFTCST DTKNVQFVFD AVTDVIIKNN LKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 37.915496 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD ...String: MGSLLQSELD QLRQEAEQLK NQIRDARKAC ADATLSQITN NIDPVGRIQM RTRRTLRGHL AKIYAMHWGT DSRLLVSASQ DGKLIIWDS YTTNKVHAIP LRSSWVMTCA YAPSGNYVAC GGLDNICSIY NLKTREGNVR VSRELAGHTG YLSCCRFLDD N QIVTSSGD TTCALWDIET GQQTTTFTGH TGDVMSLSLA PDTRLFVSGA CDASAKLWDV REGMCRQTFT GHESDINAIC FF PNGNAFA TGSDDATCRL FDLRADQELM TYSHDNIICG ITSVSFSKSG RLLLAGYDDF NCNVWDALKA DRAGVLAGHD NRV SCLGVT DDGMAVATGS WDSFLKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-Macromolecule #5: Guanine nucleotide-binding protein subunit gamma
| Macromolecule | Name: Guanine nucleotide-binding protein subunit gamma / type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 7.432554 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: ASNNTASIAQ ARKLVEQLKM EANIDRIKVS KAAADLMAYC EAHAKEDPLL TPVPASENPF REKKFFC UniProtKB: Guanine nucleotide-binding protein subunit gamma |
-Macromolecule #6: PALMITIC ACID
| Macromolecule | Name: PALMITIC ACID / type: ligand / ID: 6 / Number of copies: 2 / Formula: PLM |
|---|---|
| Molecular weight | Theoretical: 256.424 Da |
| Chemical component information |  ChemComp-PLM: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.2 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 5.0 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller
































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)