[English] 日本語
 Yorodumi
Yorodumi- EMDB-34726: The structure of LcnA, LciA, and the man-PTS of Lactococcus lactis -
+ Open data
Open data
- Basic information
Basic information
| Entry | 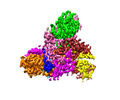 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
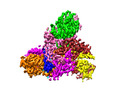
| Title | The structure of LcnA, LciA, and the man-PTS of Lactococcus lactis | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | antibiotic resistance / antimicrobial peptides / mannose phosphotransferase system / man-PTS / bacteriocins / non-pediocin-like/class IId bacteriocins / lactococcin A / LcnA / immunity / LciA / self-protection / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationprotein-Npi-phosphohistidine-sugar phosphotransferase / protein-N(pi)-phosphohistidine--N-acetyl-D-glucosamine phosphotransferase activity / bacteriocin immunity / cytolysis / phosphoenolpyruvate-dependent sugar phosphotransferase system / defense response to bacterium / host cell plasma membrane / extracellular region / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Lactococcus lactis subsp. lactis (strain KF147) (lactic acid bacteria) / Lactococcus lactis subsp. lactis (strain KF147) (lactic acid bacteria) /  Lactococcus lactis subsp. lactis (lactic acid bacteria) Lactococcus lactis subsp. lactis (lactic acid bacteria) | |||||||||
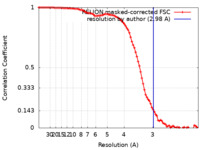
| Method | single particle reconstruction / cryo EM / Resolution: 2.98 Å | |||||||||
 Authors Authors | Wang JW | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Appl Environ Microbiol / Year: 2023 Journal: Appl Environ Microbiol / Year: 2023Title: Structural Basis of the Mechanisms of Action and Immunity of Lactococcin A, a Class IId Bacteriocin. Authors: Ruilian Li / Jinsong Duan / Yicheng Zhou / Jiawei Wang /   Abstract: Lactococcin A (LcnA), a class IId bacteriocin, induces membrane leakage and cell death by specifically binding to the membrane receptor-mannose phosphotransferase system (man-PTS), as is the case for ...Lactococcin A (LcnA), a class IId bacteriocin, induces membrane leakage and cell death by specifically binding to the membrane receptor-mannose phosphotransferase system (man-PTS), as is the case for pediocin-like (class IIa) bacteriocins. The cognate immunity protein of bacteriocins, which protects the producer cell from its own bacteriocin, recognizes and binds to the bacteriocin-man-PTS complex, consequently blocking membrane leakage. We previously deciphered the mode of action and immunity of class IIa bacteriocins. Here, we determined the structure of the ternary complex of LcnA, LciA (, the immunity protein), and its receptor, , the man-PTS of Lactococcus lactis (ll-man-PTS). An external loop on the membrane-located component IIC of ll-man-PTS was found to prevent specific binding of the N-terminal region of LcnA to the site recognized by pediocin-like bacteriocins. Thus, the N-terminal β-sheet region of LcnA recognized an adjacent site on the extracellular side of ll-man-PTS, with the LcnA C-terminal hydrophobic helix penetrating into the membrane. The cytoplasmic cleft formed within the man-PTS Core and Vmotif domains induced by embedded LcnA from the periplasmic side is adopted by the appropriate angle between helices H3 and H4 of the N terminus of LciA. The flexible C terminus of LciA then blocks membrane leakage. To summarize, our findings reveal the molecular mechanisms of action and immunity of LcnA and LciA, laying a foundation for further design of class IId bacteriocins. Class IId (lactococcin-like) bacteriocins and class IIa (pediocin-like) bacteriocins share a few similarities: (i) both induce membrane leakage and cell death by specifically binding the mannose phosphotransferase system (man-PTS) on their target cells, and (ii) cognate immunity proteins recognize and bind to the bacteriocin-man-PTS complex to block membrane leakage. However, class IId bacteriocins lack the "pediocin box" motif, which is typical of class IIa bacteriocins, and basically target only lactococcal cells; in contrast, class IIa bacteriocins target diverse bacterial cells, but not lactococcal cells. We previously solved the structure of class IIa bacteriocin-receptor-immunity ternary complex from Lactobacillus sakei. Here, we determined the structure of the ternary complex of class IId bacteriocin LcnA, its cognate immunity protein LciA, and its receptor, the man-PTS of Lactococcus lactis. By comparing the interactions between man-PTS and class IIa and class IId bacteriocins, this study affords some clues to better understand the specificity of bacteriocins targeting the mannose phosphotransferase system. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34726.map.gz emd_34726.map.gz | 6.5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34726-v30.xml emd-34726-v30.xml emd-34726.xml emd-34726.xml | 17.7 KB 17.7 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34726_fsc.xml emd_34726_fsc.xml | 9.1 KB | Display |  FSC data file FSC data file |
| Images |  emd_34726.png emd_34726.png | 92.3 KB | ||
| Filedesc metadata |  emd-34726.cif.gz emd-34726.cif.gz | 5.9 KB | ||
| Others |  emd_34726_half_map_1.map.gz emd_34726_half_map_1.map.gz emd_34726_half_map_2.map.gz emd_34726_half_map_2.map.gz | 49.5 MB 49.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34726 http://ftp.pdbj.org/pub/emdb/structures/EMD-34726 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34726 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34726 | HTTPS FTP |
-Validation report
| Summary document |  emd_34726_validation.pdf.gz emd_34726_validation.pdf.gz | 665.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34726_full_validation.pdf.gz emd_34726_full_validation.pdf.gz | 665.5 KB | Display | |
| Data in XML |  emd_34726_validation.xml.gz emd_34726_validation.xml.gz | 14.7 KB | Display | |
| Data in CIF |  emd_34726_validation.cif.gz emd_34726_validation.cif.gz | 20.3 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34726 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34726 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34726 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34726 | HTTPS FTP |
-Related structure data
| Related structure data |  8hfsMC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34726.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34726.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.0742 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: half map 1
| File | emd_34726_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 1 | ||||||||||||
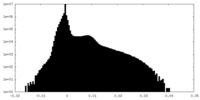
| Projections & Slices |
| ||||||||||||
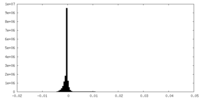
| Density Histograms |
-Half map: half map 2
| File | emd_34726_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | half map 2 | ||||||||||||
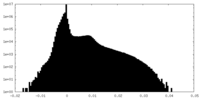
| Projections & Slices |
| ||||||||||||
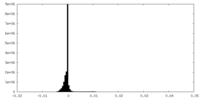
| Density Histograms |
- Sample components
Sample components
-Entire : The structure of LcnA, LciA, and the man-PTS of Lactococcus lactis
| Entire | Name: The structure of LcnA, LciA, and the man-PTS of Lactococcus lactis |
|---|---|
| Components |
|
-Supramolecule #1: The structure of LcnA, LciA, and the man-PTS of Lactococcus lactis
| Supramolecule | Name: The structure of LcnA, LciA, and the man-PTS of Lactococcus lactis type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#4 |
|---|---|
| Source (natural) | Organism:  Lactococcus lactis subsp. lactis (strain KF147) (lactic acid bacteria) Lactococcus lactis subsp. lactis (strain KF147) (lactic acid bacteria) |
-Macromolecule #1: Mannose-specific PTS system, IIC component
| Macromolecule | Name: Mannose-specific PTS system, IIC component / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO EC number: protein-Npi-phosphohistidine-sugar phosphotransferase |
|---|---|
| Source (natural) | Organism:  Lactococcus lactis subsp. lactis (strain KF147) (lactic acid bacteria) Lactococcus lactis subsp. lactis (strain KF147) (lactic acid bacteria) |
| Molecular weight | Theoretical: 27.572371 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MEYGVLSVIL VIVVAFLAGL EGILDQWQFH QPIIACSLIG IVTGHASAGI ILGGSLQLIA LGWANVGAAV APDAALASIA SSILMVQSN NFDLTHIMGT IVPAAILLAT AGLVLTTLVR MLSVVLVHQA DRAAENGSYS GVEMWHFIAL ICQGLRIAIP A GLLLVISP ...String: MEYGVLSVIL VIVVAFLAGL EGILDQWQFH QPIIACSLIG IVTGHASAGI ILGGSLQLIA LGWANVGAAV APDAALASIA SSILMVQSN NFDLTHIMGT IVPAAILLAT AGLVLTTLVR MLSVVLVHQA DRAAENGSYS GVEMWHFIAL ICQGLRIAIP A GLLLVISP DAIQKALAAI PPVISGGLAV GGGMVVAVGY AMVINLMATR EVWPFFFLGF ALAPISELTL IATGVLGVVI AI VYLNLQA SGGSGNGTAS SSGDPIGDIL NDY UniProtKB: Mannose-specific PTS system, IIC component |
-Macromolecule #2: Mannose-specific PTS system, IID component
| Macromolecule | Name: Mannose-specific PTS system, IID component / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO EC number: protein-Npi-phosphohistidine-sugar phosphotransferase |
|---|---|
| Source (natural) | Organism:  Lactococcus lactis subsp. lactis (strain KF147) (lactic acid bacteria) Lactococcus lactis subsp. lactis (strain KF147) (lactic acid bacteria) |
| Molecular weight | Theoretical: 33.630473 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MSENKVTLDK KIRRSVMWRS MFLQGSWNYE RMQNGGWAYS LIPALKKLYP SGEEAKEALK RHLEFFNTHP YVAAPIIGVT LALEEERAN GADIDDAAIQ GVKVGMMGPL AGIGDPVFWF TVRPIVGAIA ASLATGGSII APLFFFIVWN AIRIAFLWYT Q EFGYKSGS ...String: MSENKVTLDK KIRRSVMWRS MFLQGSWNYE RMQNGGWAYS LIPALKKLYP SGEEAKEALK RHLEFFNTHP YVAAPIIGVT LALEEERAN GADIDDAAIQ GVKVGMMGPL AGIGDPVFWF TVRPIVGAIA ASLATGGSII APLFFFIVWN AIRIAFLWYT Q EFGYKSGS AITKDLGGGL LQTVTKGASI LGMFVLGVLI QRWVTINFNG PNAVVSKIPL QKGAYVEFPK GSVSGTQLHD IL GQVGNKL SLDPTKVTYL QDNLNQLIPG LAGLLITLLC MWLLKKKVSP IVIIFGLFVV GILGRWAQIM UniProtKB: Mannose-specific PTS system, IID component |
-Macromolecule #3: Lactococcin-A immunity protein
| Macromolecule | Name: Lactococcin-A immunity protein / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Lactococcus lactis subsp. lactis (lactic acid bacteria) Lactococcus lactis subsp. lactis (lactic acid bacteria) |
| Molecular weight | Theoretical: 11.179947 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKKKQIEFEN ELRSMLATAL EKDISQEERN ALNIAEKALD NSEYLPKIIL NLRKALTPLA INRTLNHDLS ELYKFITSSK ASNKNLGGG LIMSWGRLF UniProtKB: Lactococcin-A immunity protein |
-Macromolecule #4: Bacteriocin lactococcin-A
| Macromolecule | Name: Bacteriocin lactococcin-A / type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Lactococcus lactis subsp. lactis (lactic acid bacteria) Lactococcus lactis subsp. lactis (lactic acid bacteria) |
| Molecular weight | Theoretical: 5.783341 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: KLTFIQSTAA GDLYYNTNTH KYVYQQTQNA FGAAANTIVN GWMGGAAGGF GLHH UniProtKB: Bacteriocin lactococcin-A |
-Macromolecule #5: alpha-D-mannopyranose
| Macromolecule | Name: alpha-D-mannopyranose / type: ligand / ID: 5 / Number of copies: 3 / Formula: MAN |
|---|---|
| Molecular weight | Theoretical: 180.156 Da |
| Chemical component information |  ChemComp-MAN: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Refinement | Space: REAL |
|---|---|
| Output model |  PDB-8hfs: |
 Movie
Movie Controller
Controller


 Z
Z Y
Y X
X