+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Icosahedral reconstruction of HCMV A-capsid | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | A-capsid / icosahedral recontruction / VIRUS | |||||||||
| Biological species |   Human betaherpesvirus 5 Human betaherpesvirus 5 | |||||||||
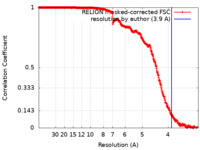
| Method | single particle reconstruction / cryo EM / Resolution: 3.9 Å | |||||||||
 Authors Authors | Li Z / Yu X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Cryo-electron microscopy structures of capsids and in situ portals of DNA-devoid capsids of human cytomegalovirus. Authors: Zhihai Li / Jingjing Pang / Rongchao Gao / Qingxia Wang / Maoyan Zhang / Xuekui Yu /  Abstract: The portal-scaffold complex is believed to nucleate the assembly of herpesvirus procapsids. During capsid maturation, two events occur: scaffold expulsion and DNA incorporation. The portal-scaffold ...The portal-scaffold complex is believed to nucleate the assembly of herpesvirus procapsids. During capsid maturation, two events occur: scaffold expulsion and DNA incorporation. The portal-scaffold interaction and the conformational changes that occur to the portal during the different stages of capsid formation have yet to be elucidated structurally. Here we present high-resolution structures of the A- and B-capsids and in-situ portals of human cytomegalovirus. We show that scaffolds bind to the hydrophobic cavities formed by the dimerization and Johnson-fold domains of the major capsid proteins. We further show that 12 loop-helix-loop fragments-presumably from the scaffold domain-insert into the hydrophobic pocket of the portal crown domain. The portal also undergoes significant changes both positionally and conformationally as it accompanies DNA packaging. These findings unravel the mechanism by which the portal interacts with the scaffold to nucleate capsid assembly and further our understanding of scaffold expulsion and DNA incorporation. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34698.map.gz emd_34698.map.gz | 3.8 GB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34698-v30.xml emd-34698-v30.xml emd-34698.xml emd-34698.xml | 11.6 KB 11.6 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_34698_fsc.xml emd_34698_fsc.xml | 35.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_34698.png emd_34698.png | 63.4 KB | ||
| Masks |  emd_34698_msk_1.map emd_34698_msk_1.map | 4 GB |  Mask map Mask map | |
| Filedesc metadata |  emd-34698.cif.gz emd-34698.cif.gz | 3.9 KB | ||
| Others |  emd_34698_half_map_1.map.gz emd_34698_half_map_1.map.gz emd_34698_half_map_2.map.gz emd_34698_half_map_2.map.gz | 3.3 GB 3.3 GB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34698 http://ftp.pdbj.org/pub/emdb/structures/EMD-34698 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34698 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34698 | HTTPS FTP |
-Validation report
| Summary document |  emd_34698_validation.pdf.gz emd_34698_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34698_full_validation.pdf.gz emd_34698_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_34698_validation.xml.gz emd_34698_validation.xml.gz | 42.5 KB | Display | |
| Data in CIF |  emd_34698_validation.cif.gz emd_34698_validation.cif.gz | 55.9 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34698 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34698 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34698 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34698 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_34698.map.gz / Format: CCP4 / Size: 4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34698.map.gz / Format: CCP4 / Size: 4 GB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.625 Å | ||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_34698_msk_1.map emd_34698_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #2
| File | emd_34698_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: #1
| File | emd_34698_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Human betaherpesvirus 5
| Entire | Name:   Human betaherpesvirus 5 Human betaherpesvirus 5 |
|---|---|
| Components |
|
-Supramolecule #1: Human betaherpesvirus 5
| Supramolecule | Name: Human betaherpesvirus 5 / type: virus / ID: 1 / Parent: 0 / NCBI-ID: 10359 / Sci species name: Human betaherpesvirus 5 / Virus type: VIRION / Virus isolate: STRAIN / Virus enveloped: No / Virus empty: Yes |
|---|
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 30.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 0.9 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller






















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)