[English] 日本語
 Yorodumi
Yorodumi- EMDB-34407: SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | SARS-CoV-2 Omicron BA.1 Spike glycoprotein in complex with rabbit monoclonal antibody 1H1 Fab in the class 1 conformation | |||||||||
 Map data Map data | ||||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | VIRAL PROTEIN-IMMUNE SYSTEM COMPLEX | |||||||||
| Function / homology |  Function and homology information Function and homology informationMaturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion ...Maturation of spike protein / Translation of Structural Proteins / Virion Assembly and Release / host cell surface / host extracellular space / viral translation / symbiont-mediated-mediated suppression of host tetherin activity / Induction of Cell-Cell Fusion / structural constituent of virion / membrane fusion / entry receptor-mediated virion attachment to host cell / Attachment and Entry / host cell endoplasmic reticulum-Golgi intermediate compartment membrane / positive regulation of viral entry into host cell / receptor-mediated virion attachment to host cell / host cell surface receptor binding / symbiont-mediated suppression of host innate immune response / receptor ligand activity / endocytosis involved in viral entry into host cell / fusion of virus membrane with host plasma membrane / fusion of virus membrane with host endosome membrane / viral envelope / virion attachment to host cell / SARS-CoV-2 activates/modulates innate and adaptive immune responses / host cell plasma membrane / virion membrane / identical protein binding / membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |   | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 3.41 Å | |||||||||
 Authors Authors | Guo H / Gao Y / Lu Y / Yang H / Ji X | |||||||||
| Funding support |  China, 1 items China, 1 items
| |||||||||
 Citation Citation |  Journal: Commun Biol / Year: 2023 Journal: Commun Biol / Year: 2023Title: Mechanism of a rabbit monoclonal antibody broadly neutralizing SARS-CoV-2 variants. Authors: Hangtian Guo / Yixuan Yang / Tiantian Zhao / Yuchi Lu / Yan Gao / Tinghan Li / Hang Xiao / Xiaoyu Chu / Le Zheng / Wanting Li / Hao Cheng / Haibin Huang / Yang Liu / Yang Lou / Henry C ...Authors: Hangtian Guo / Yixuan Yang / Tiantian Zhao / Yuchi Lu / Yan Gao / Tinghan Li / Hang Xiao / Xiaoyu Chu / Le Zheng / Wanting Li / Hao Cheng / Haibin Huang / Yang Liu / Yang Lou / Henry C Nguyen / Chao Wu / Yuxin Chen / Haitao Yang / Xiaoyun Ji /  Abstract: Due to the continuous evolution of SARS-CoV-2, the Omicron variant has emerged and exhibits severe immune evasion. The high number of mutations at key antigenic sites on the spike protein has made a ...Due to the continuous evolution of SARS-CoV-2, the Omicron variant has emerged and exhibits severe immune evasion. The high number of mutations at key antigenic sites on the spike protein has made a large number of existing antibodies and vaccines ineffective against this variant. Therefore, it is urgent to develop efficient broad-spectrum neutralizing therapeutic drugs. Here we characterize a rabbit monoclonal antibody (RmAb) 1H1 with broad-spectrum neutralizing potency against Omicron sublineages including BA.1, BA.1.1, BA.2, BA.2.12.1, BA.2.75, BA.3 and BA.4/5. Cryo-electron microscopy (cryo-EM) structure determination of the BA.1 spike-1H1 Fab complexes shows that 1H1 targets a highly conserved region of RBD and avoids most of the circulating Omicron mutations, explaining its broad-spectrum neutralization potency. Our findings indicate 1H1 as a promising RmAb model for designing broad-spectrum neutralizing antibodies and shed light on the development of therapeutic agents as well as effective vaccines against newly emerging variants in the future. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_34407.map.gz emd_34407.map.gz | 483.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-34407-v30.xml emd-34407-v30.xml emd-34407.xml emd-34407.xml | 21.6 KB 21.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_34407.png emd_34407.png | 51.9 KB | ||
| Filedesc metadata |  emd-34407.cif.gz emd-34407.cif.gz | 7.3 KB | ||
| Others |  emd_34407_half_map_1.map.gz emd_34407_half_map_1.map.gz emd_34407_half_map_2.map.gz emd_34407_half_map_2.map.gz | 475.3 MB 475.3 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-34407 http://ftp.pdbj.org/pub/emdb/structures/EMD-34407 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34407 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-34407 | HTTPS FTP |
-Validation report
| Summary document |  emd_34407_validation.pdf.gz emd_34407_validation.pdf.gz | 1 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_34407_full_validation.pdf.gz emd_34407_full_validation.pdf.gz | 1 MB | Display | |
| Data in XML |  emd_34407_validation.xml.gz emd_34407_validation.xml.gz | 19 KB | Display | |
| Data in CIF |  emd_34407_validation.cif.gz emd_34407_validation.cif.gz | 22.7 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34407 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34407 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34407 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-34407 | HTTPS FTP |
-Related structure data
| Related structure data |  8h00MC  8gzzC  8h01C  8ituC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_34407.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_34407.map.gz / Format: CCP4 / Size: 512 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.832 Å | ||||||||||||||||||||||||||||||||||||
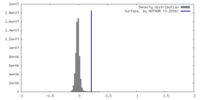
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_34407_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
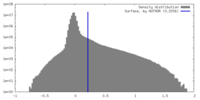
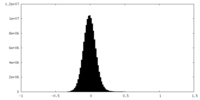
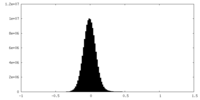
| Density Histograms |
-Half map: #1
| File | emd_34407_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
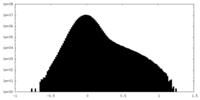
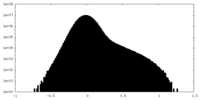
| Density Histograms |
- Sample components
Sample components
-Entire : SARS-CoV-2 Omicron spike glycoprotein complex with 55A8 Fab
| Entire | Name: SARS-CoV-2 Omicron spike glycoprotein complex with 55A8 Fab |
|---|---|
| Components |
|
-Supramolecule #1: SARS-CoV-2 Omicron spike glycoprotein complex with 55A8 Fab
| Supramolecule | Name: SARS-CoV-2 Omicron spike glycoprotein complex with 55A8 Fab type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #2: Omicron spike glycoprotein
| Supramolecule | Name: Omicron spike glycoprotein / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1 Details: SARS-CoV-2 Omicron BA.1 variant spike protein ectodomain |
|---|---|
| Source (natural) | Organism:  |
-Supramolecule #3: 1H1 Fab
| Supramolecule | Name: 1H1 Fab / type: complex / ID: 3 / Parent: 1 / Macromolecule list: #2-#3 Details: Fab fragment generated by proteolytic cleavage of IgG antibody |
|---|
-Macromolecule #1: Spike glycoprotein
| Macromolecule | Name: Spike glycoprotein / type: protein_or_peptide / ID: 1 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 142.620094 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHVISG TNGTKRFDNP VLPFNDGVY FASIEKSNII RGWIFGTTLD SKTQSLLIVN NATNVVIKVC EFQFCNDPFL DHKNNKSWME SEFRVYSSAN N CTFEYVSQ ...String: MFVFLVLLPL VSSQCVNLTT RTQLPPAYTN SFTRGVYYPD KVFRSSVLHS TQDLFLPFFS NVTWFHVISG TNGTKRFDNP VLPFNDGVY FASIEKSNII RGWIFGTTLD SKTQSLLIVN NATNVVIKVC EFQFCNDPFL DHKNNKSWME SEFRVYSSAN N CTFEYVSQ PFLMDLEGKQ GNFKNLREFV FKNIDGYFKI YSKHTPINIV REPEDLPQGF SALEPLVDLP IGINITRFQT LL ALHRSYL TPGDSSSGWT AGAAAYYVGY LQPRTFLLKY NENGTITDAV DCALDPLSET KCTLKSFTVE KGIYQTSNFR VQP TESIVR FPNITNLCPF DEVFNATRFA SVYAWNRKRI SNCVADYSVL YNLAPFFTFK CYGVSPTKLN DLCFTNVYAD SFVI RGDEV RQIAPGQTGN IADYNYKLPD DFTGCVIAWN SNKLDSKVSG NYNYLYRLFR KSNLKPFERD ISTEIYQAGN KPCNG VAGF NCYFPLRSYS FRPTYGVGHQ PYRVVVLSFE LLHAPATVCG PKKSTNLVKN KCVNFNFNGL KGTGVLTESN KKFLPF QQF GRDIADTTDA VRDPQTLEIL DITPCSFGGV SVITPGTNTS NQVAVLYQGV NCTEVPVAIH ADQLTPTWRV YSTGSNV FQ TRAGCLIGAE YVNNSYECDI PIGAGICASY QTQTKSHGSA SSVASQSIIA YTMSLGAENS VAYSNNSIAI PTNFTISV T TEILPVSMTK TSVDCTMYIC GDSTECSNLL LQYGSFCTQL KRALTGIAVE QDKNTQEVFA QVKQIYKTPP IKYFGGFNF SQILPDPSKP SKRSPIEDLL FNKVTLADAG FIKQYGDCLG DIAARDLICA QKFKGLTVLP PLLTDEMIAQ YTSALLAGTI TSGWTFGAG PALQIPFPMQ MAYRFNGIGV TQNVLYENQK LIANQFNSAI GKIQDSLSST PSALGKLQDV VNHNAQALNT L VKQLSSKF GAISSVLNDI FSRLDPPEAE VQIDRLITGR LQSLQTYVTQ QLIRAAEIRA SANLAATKMS ECVLGQSKRV DF CGKGYHL MSFPQSAPHG VVFLHVTYVP AQEKNFTTAP AICHDGKAHF PREGVFVSNG THWFVTQRNF YEPQIITTDN TFV SGNCDV VIGIVNNTVY DPLQPELDSF KEELDKYFKN HTSPDVDLGD ISGINASVVN IQKEIDRLNE VAKNLNESLI DLQE LGKYE QGSGYIPEAP RDGQAYVRKD GEWVFLSTFL SGLEVLFQGP GGWSHPQFEK GGGSGGGSGG SAWSHPQFEK GGSHH HHHH HH UniProtKB: Spike glycoprotein |
-Macromolecule #2: rabbit monoclonal antibody 1H1 Fab light chain
| Macromolecule | Name: rabbit monoclonal antibody 1H1 Fab light chain / type: protein_or_peptide / ID: 2 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 11.739082 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: DIVMTQTPAS VSEPVGGTVT IKCQASESIS NWLAWYQQKP GQPPKLLIYA AFTLASGVPS RFKGSGSGTQ FTLTINGVEC ADAATYYCQ QTYSSRDVDN VFGGGTEVVV KG |
-Macromolecule #3: rabbit monoclonal antibody 1H1 Fab heavy chain
| Macromolecule | Name: rabbit monoclonal antibody 1H1 Fab heavy chain / type: protein_or_peptide / ID: 3 / Number of copies: 3 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 12.863201 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: QSLEESGGDL VKPGASLTLT CTASGFSFSS GYDMCWVRQA PGKGLEWIAC IGTGSSGNIY YASWAKGRFT ISKTSSTTVT LQMTSLTAA DTATYFCARD DADYAGPDYF NLWGPGTLVT VSS |
-Macromolecule #5: 2-acetamido-2-deoxy-beta-D-glucopyranose
| Macromolecule | Name: 2-acetamido-2-deoxy-beta-D-glucopyranose / type: ligand / ID: 5 / Number of copies: 15 / Formula: NAG |
|---|---|
| Molecular weight | Theoretical: 221.208 Da |
| Chemical component information |  ChemComp-NAG: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.8000000000000003 µm / Nominal defocus min: 1.2 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)