+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3413 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | cryo-EM of nanoscale DNA assemblies | |||||||||
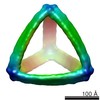
 Map data Map data | Reconstruction of a DNA nested cube | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | Scaffolded DNA origami | |||||||||
| Biological species |  Enterobacteria phage M13 (virus) / synthetic construct (others) Enterobacteria phage M13 (virus) / synthetic construct (others) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 40.0 Å | |||||||||
 Authors Authors | Zhang K / Chiu W / Veneziano R / Ratanalert S / Zhang F / Yan H / Bathe M | |||||||||
 Citation Citation |  Journal: Science / Year: 2016 Journal: Science / Year: 2016Title: Designer nanoscale DNA assemblies programmed from the top down. Authors: Rémi Veneziano / Sakul Ratanalert / Kaiming Zhang / Fei Zhang / Hao Yan / Wah Chiu / Mark Bathe /  Abstract: Scaffolded DNA origami is a versatile means of synthesizing complex molecular architectures. However, the approach is limited by the need to forward-design specific Watson-Crick base pairing manually ...Scaffolded DNA origami is a versatile means of synthesizing complex molecular architectures. However, the approach is limited by the need to forward-design specific Watson-Crick base pairing manually for any given target structure. Here, we report a general, top-down strategy to design nearly arbitrary DNA architectures autonomously based only on target shape. Objects are represented as closed surfaces rendered as polyhedral networks of parallel DNA duplexes, which enables complete DNA scaffold routing with a spanning tree algorithm. The asymmetric polymerase chain reaction is applied to produce stable, monodisperse assemblies with custom scaffold length and sequence that are verified structurally in three dimensions to be high fidelity by single-particle cryo-electron microscopy. Their long-term stability in serum and low-salt buffer confirms their utility for biological as well as nonbiological applications. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3413.map.gz emd_3413.map.gz | 30.4 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3413-v30.xml emd-3413-v30.xml emd-3413.xml emd-3413.xml | 16.2 KB 16.2 KB | Display Display |  EMDB header EMDB header |
| Images |  EMD-3413.png EMD-3413.png | 158.3 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3413 http://ftp.pdbj.org/pub/emdb/structures/EMD-3413 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3413 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3413 | HTTPS FTP |
-Validation report
| Summary document |  emd_3413_validation.pdf.gz emd_3413_validation.pdf.gz | 205.9 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_3413_full_validation.pdf.gz emd_3413_full_validation.pdf.gz | 205 KB | Display | |
| Data in XML |  emd_3413_validation.xml.gz emd_3413_validation.xml.gz | 7.4 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3413 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3413 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3413 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-3413 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3413.map.gz / Format: CCP4 / Size: 210.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3413.map.gz / Format: CCP4 / Size: 210.9 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Reconstruction of a DNA nested cube | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.95 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : DNA nested cube, 73-bp edge length
| Entire | Name: DNA nested cube, 73-bp edge length |
|---|---|
| Components |
|
-Supramolecule #1000: DNA nested cube, 73-bp edge length
| Supramolecule | Name: DNA nested cube, 73-bp edge length / type: sample / ID: 1000 Details: Monodisperse and pure sample were prepared by overnight folding reaction and purified using centrifugal filter (MWCO 100KDa) Oligomeric state: monomer / Number unique components: 2 |
|---|---|
| Molecular weight | Experimental: 2.06 MDa / Theoretical: 2.06 MDa / Method: Calculation |
-Macromolecule #1: M13mp18 phage genome segment
| Macromolecule | Name: M13mp18 phage genome segment / type: dna / ID: 1 / Name.synonym: Scaffold strand / Details: amplified using assymetric PCR / Classification: DNA / Structure: SINGLE STRANDED / Synthetic?: No |
|---|---|
| Source (natural) | Organism:  Enterobacteria phage M13 (virus) / Strain: M13mp18 / synonym: M13 phage Enterobacteria phage M13 (virus) / Strain: M13mp18 / synonym: M13 phage |
| Molecular weight | Experimental: 1.03 MDa / Theoretical: 1.03 MDa |
| Sequence | String: GTCTCGCTGG TGAAAAGAAA AACCACCCTG GCGCCCAATA CGCAAACCGC CTCTCCCCGC GCGTTGGCCG ATTCATTAAT GCAGCTGGCA CGACAGGTTT CCCGACTGGA AAGCGGGCAG TGAGCGCAAC GCAATTAATG TGAGTTAGCT CACTCATTAG GCACCCCAGG ...String: GTCTCGCTGG TGAAAAGAAA AACCACCCTG GCGCCCAATA CGCAAACCGC CTCTCCCCGC GCGTTGGCCG ATTCATTAAT GCAGCTGGCA CGACAGGTTT CCCGACTGGA AAGCGGGCAG TGAGCGCAAC GCAATTAATG TGAGTTAGCT CACTCATTAG GCACCCCAGG CTTTACACTT TATGCTTCCG GCTCGTATGT TGTGTGGAAT TGTGAGCGGA TAACAATTTC ACACAGGAAA CAGCTATGAC CATGATTACG AATTCGAGCT CGGTACCCGG GGATCCTCTA GAGTCGACCT GCAGGCATGC AAGCTTGGCA CTGGCCGTCG TTTTACAACG TCGTGACTGG GAAAACCCTG GCGTTACCCA ACTTAATCGC CTTGCAGCAC ATCCCCCTTT CGCCAGCTGG CGTAATAGCG AAGAGGCCCG CACCGATCGC CCTTCCCAAC AGTTGCGCAG CCTGAATGGC GAATGGCGCT TTGCCTGGTT TCCGGCACCA GAAGCGGTGC CGGAAAGCTG GCTGGAGTGC GATCTTCCTG AGGCCGATAC GGTCGTCGTC CCCTCAAACT GGCAGATGCA CGGTTACGAT GCGCCCATCT ACACCAACGT AACCTATCCC ATTACGGTCA ATCCGCCGTT TGTTCCCACG GAGAATCCGA CGGGTTGTTA CTCGCTCACA TTTAATGTTG ATGAAAGCTG GCTACAGGAA GGCCAGACGC GAATTATTTT TGATGGCGTT CCTATTGGTT AAAAAATGAG CTGATTTAAC AAAAATTTAA CGCGAATTTT AACAAAATAT TAACGTTTAC AATTTAAATA TTTGCTTATA CAATCTTCCT GTTTTTGGGG CTTTTCTGAT TATCAACCGG GGTACATATG ATTGACATGC TAGTTTTACG ATTACCGTTC ATCGATTCTC TTGTTTGCTC CAGACTCTCA GGCAATGACC TGATAGCCTT TGTAGATCTC TCAAAAATAG CTACCCTCTC CGGCATTAAT TTATCAGCTA GAACGGTTGA ATATCATATT GATGGTGATT TGACTGTCTC CGGCCTTTCT CACCCTTTTG AATCTTTACC TACACATTAC TCAGGCATTG CATTTAAAAT ATATGAGGGT TCTAAAAATT TTTATCCTTG CGTTGAAATA AAGGCTTCTC CCGCAAAAGT ATTACAGGGT CATAATGTTT TTGGTACAAC CGATTTAGCT TTATGCTCTG AGGCTTTATT GCTTAATTTT GCTAATTCTT TGCCTTGCCT GTATGATTTA TTGGATGTTA ATGCTACTAC TATTAGTAGA ATTGATGCCA CCTTTTCAGC TCGCGCCCCA AATGAAAATA TAGCTAAACA GGTTATTGAC CATTTGCGAA ATGTATCTAA TGGTCAAACT AAATCTACTC GTTCGCAGAA TTGGGAATCA ACTGTTACAT GGAATGAAAC TTCCAGACAC CGTACTTTAG TTGCATATTT AAAACATGTT GAGCTACAGC ACCAGATTCA GCAATTAAGC TCTAAGCCAT CCGCAAAAAT GACCTCTTAT CAAAAGGAGC AATTAAAGGT ACTCTCTAAT CCTGACCTGT TGGAGTTTGC TTCCGGTCTG GTTCGCTTTG AAGCTCGAAT TAAAACGCGA TATTTGAAGT CTTTCGGGCT TCCTCTTAAT CTTTTTGATG CAATCCGCTT TGCTTCTGAC TATAATAGTC AGGGTAAAGA CCTGATTTTT GATTTATGGT CATTCTCGTT TTCTGAACTG TTTAAAGCAT TTGAGGGGGA TTCAATGAAT ATTTATGACG ATTCCGCAGT ATTGGACGCT ATCCAGTCTA AACATTTTAC TATTACCCCC TCTGGCAAAA CTTCTTTTGC AAAAGCCTCT CGCTATTTTG GTTTTTATCG TCGTCTGGTA AACGAGGGTT ATGATAGTGT TGCTCTTACT ATGCCTCGTA ATTCCTTTTG GCGTTATGTA TCTGCATTAG TTGAATGTGG TATTCCTAAA TCTCAACTGA TGAATCTTTC TACCTGTAAT AATGTTGTTC CGTTAGTTCG TTTTATTAAC GTAGATTTTT CTTCCCAACG TCCTGACTGG TATAATGAGC CAGTTCTTAA AATCGCATAA GGTAATTCAC AATGATTAAA GTTGAAATTA AACCATCTCA AGCCCAATTT ACTACTCGTT CTGGTGTTCT CGTCAGGGCA AGCCTTATTC ACTGAATGAG CAGCTTTGTT ACGTTGATTT GGGTAATGAA TATCCGGTTC TTGTCAAGAT TACTCTTGAT GAAGGTCAGC CAGCCTATGC GCCTGGTCTG TACACCGTTC ATCTGTCCTC TTTCAAAGTT GGTCAGTTCG GTTCCCTTAT GATTGACCGT CTGCGCCTCG TTCCGGCTAA GTAACATGGA GCAGGTCGCG GATTTCGACA CAATTTATCA GGCGATGATA CAAATCTCCG TTGTACTTTG TTTCGCGCTT GGTATAATCG CTGGGGGTCA AAGATGAGTG TTTTAGTGTA TTCTTTCGCC TCTTTCGTTT TAGGTTGGTG CCTTCGTAGT GGCATTACGT ATTTTACCCG TTTAATGGAA ACTTCCTCAT GAAAAAGTCT TTAGTCCTCA AAGCCTCTGT AGCCGTTGCT ACCCTCGTTC CGATGCTGTC TTTCGCTGCT GAGGGTGACG ATCCCGCAAA AGCGGCCTTT AACTCCCTGC AAGCCTCAGC GACCGAATAT ATCGGTTATG CGTGGGCGAT GGTTGTTGTC ATTGTCGGCG CAACTATCGG TATCAAGCTG TTTAAGAAAT TCACCTCGAA AGCAAGCTGA TAAACCGATA CAATTAAAGG CTCCTTTTGG AGCCTTTTTT TTTGGAGATT TTCAACGTGA AAAAATTATT ATTCGCAATT CCTTTAGTTG TTCCTTTCTA TTCTCACTCC GCTGAAACTG TTGAAAGTTG TTTAGCAAAA CCCCATACAG AAAATTCATT TACTAACGTC TGGAAAGACG ACAAAACTTT AGATCGTTAC GCTAACTATG AGGGTTGTCT GTGGAATGCT ACAGGCGTTG TAGTTTGTAC TGGTGACGAA ACTCAGTGTT ACGGTACATG GGTTCCTATT GGGCTTGCTA TCCCTGAAAA TGAGGGTGGT GGCTCTGAGG GTGGCGGTTC TGAGGGTGGC GGTTCTGAGG GTGGCGGTAC TAAACCTCCT GAGTACGGTG ATACACCTAT TCCGGGCTAT ACTTATATCA ACCCTCTCGA CGGCACTTAT CCGCCTGGTA CTGAGCAAAA CCCCGCTAAT CCTAATCCTT CTCTTGAGGA GTCTCAGCCT CTTAATACTT TCATGTTTCA GAATAATAGG TTCCGAAATA GGCAGGGGGC ATTAAC |
-Macromolecule #2: Synthetic DNA oligonucleotides
| Macromolecule | Name: Synthetic DNA oligonucleotides / type: dna / ID: 2 / Name.synonym: Staple strand Details: AAGTTTTGCCAACGACGTTGTA GCTTTTTTTTCCGGCACCGCAACGCCAGGGTTTTTTTTTCCCAGTCGAGGGG TGTGCTTTTTTGCAAGGCGAGAAACCAGGCATTTTTAAGCGCCATTGCTGCG TTCTGGTGCCGTTAAGTTGGGT TCGGATTCTCCAGATCGCACTC ...Details: AAGTTTTGCCAACGACGTTGTA GCTTTTTTTTCCGGCACCGCAACGCCAGGGTTTTTTTTTCCCAGTCGAGGGG TGTGCTTTTTTGCAAGGCGAGAAACCAGGCATTTTTAAGCGCCATTGCTGCG TTCTGGTGCCGTTAAGTTGGGT TCGGATTCTCCAGATCGCACTC GTGCATTTTTTCTGCCAGTTACGACAGTATCTTTTTGGCCTCAGGAGTGGGA TGAGGTAGATTAACGAGGGACG TCGGGTTTTTAAACCTGTCGTAATGAATCGGTTTTTCCAACGCGCGGTTTGC TGGGCCTGCATTGCCAGGCATC ACAGATTTTTTGAACGGTGTCATAGGCTGGCTTTTTTGACCTTCATCTTGAC ACTGCCAGGCGACAGACCCGCT GACCATTTTTTTAGATACATCAATAACCTGTTTTTTTTAGCTATATAAGTAC CCAACAATGGTTTCGCATTTGA CGGCCTTTTTAGTGCCAAGCAAAGCCTGGGGTTTTTTGCCTAATGATAGCAT ACATATTTTTCGAGCCGGAACAGGTCGACTCTTTTTTAGAGGATCCAGCTCG TTGCATGCCTGGCATAAAGTGT TTTGCTTTTTGGGATCGTCAACATTCAACTATTTTTATGCAGATACTCGTTT TAAAATTTTTCGAACTAACGCTTTGAGGACTTTTTTAAAGACTTTTGGGCTT GAAAGACAGCCAGTTGAGATTTAGGAATACCCCCTCAGCAGC GGTAGCAACGATTACAGGTAGAAAGATTCATATCGGAACGAG GCTACAGAGGGAACAACATT CAAAGCGAAATATCGCGTTTTAATGTCAGGAC GTTGGGAACTGGCTCATTATACCATCGAGCTT GAAAAATCTAATTTTAAGAA ACCAGACCGGAAAGACTTCA TGACCTTTTTATAAATCAAACCATCAATATGTTTTTATATTCAACCCCTTCC TGCAATTTTTTGCCTGAGTAAAAAGATTAAGTTTTTAGGAAGCCCGAAGCAA TACCCTGACTAGGCCGGAGACAGTCAAATCAAATCAGGTCTT AAGCAAAGCGAAAGATTCAAAAGGGTGAGAAATTATAGTCAG GATTGCATCAATGTGTAGGT GTAATTTTTTACTTTTGCGGTGTAGCTCAACTTTTTATGTTTTAAAGGCGCG TTTTTGCGGAATTTTTAGAACCCTCATATATATAAGAGGTCA TTAATTGCTGTATTTCAACGCAAGGATAAAATGGCTTAGAGC AATATAATGCGAGAAGCCTT CTATCTTTTTAGGTCATTGCCATGTCAATCATTTTTTATGTACCCCTAAATT TTGTACCACCTCAGAGCATAAAGCTCGATGAA CGGTAATCGGAGCAAACAAGAGAATAAATCGG GTAAAACTAGCTGAGAGTCT AAAACATTATAGCAATAAAG CCTAATTTTTAACGAAAGAGCGCGAAACAAATTTTTGTACAACGGATTACTT CCCAGCGAACACTAAAACACTCATTTAAACAG CTTGATACTTTCGAGGTGAATTTCCTTTGACC CGATAGTTGCATCAGCTTGC TTATACCAAGGCAAAAGAAT AGGCTTGCCCTACGTAATGCCACTACGAAGGTTCAGTGAATA ACCAGAACGAGTTTCCATTAAACGGGTAAAATGACGAGAAAC GTAGTAAATTTCATGAGGAA AATGATTTTTCAACAACCATCTTTCAACAGTTTTTTTTCAGCGGAGAAGGAA CGTTAGTAAAGAGGCTTGCAGGGAGTTAAAGGTCTTTCCAGA TATGGGATTTTAACCGATATATTCGGTCGCTTGAATTTTCTG TGCTAAACAACGCCCACGCA TCCACTTTTTAGACAGCCCTAGCGAGAGGCTTTTTTTTTGCAAAAGAAACGA ACCAGTTTTTACGACGATAAGTAACGATCTATTTTTAAGTTTTGTCGCCGCT CATAGTTAGCAAACCAAAAT TTTAAACAATTCATTGAATCCCCCTAGTAAGA GCAACACTAAGGAATTACGAGGCATCAAATGC ATCATAACCCATAACGCCAA GTTCAGAAAACGTCATAAAT GTAATTTTTTAGTAAAATGTATGTGAGCGAGTTTTTTAACAACCCGCAGCCA TGTAGTTTTTCCAGCTTTCATAGCGTCCAATTTTTTACTGCGGAATCGAGAA TTAGACTGGATCAACATTAA TAAATTAATGTCAAAAATAATTCGCGTCTGGGTTCTAGCTGA AGCTATTTTTTTTTAACCAATAGGAACGCCACCGGAGAGGGT GAGAGATCTATCAGCTCATT ACAAATTTTTCGGCGGATTGGATAGGTCACGTTTTTTTGGTGTAGAGTAACC GTAAATTTTTCGTTAATATTTCGCATTAAATTTTTTTTTTGTTAAACAAAGG ACCGTAAAATTTGTTAATGG CAACTTTTTTAAAGGAATTGAGCCTTTAATTTTTTTGTATCGGTTTGCCGAC AAAAAAGGTTTTTTCACGTTGAAAACAGGAAG ATTGTATACAGAAAAGCCCCAAAAATCTCCAA AGCAAATATTGGTTGATAAT CTCCAAAAGGCGAATAATAA GTATTTTTTTGGGCGCCAGGCACATTAATTGTTTTTCGTTGCGCTCTTCCAG GTGAGCTAACTGTGGTTGCCTG GGGAGATAGATGAGAAGGCG CAAGACAATTCCGCTCAGTAAT AATTCTTTTTGTAATCATGGTTCCTGTGTGATTTTTAATTGTTATCCACACA GAGATTTTTTGGTTTAATTTCATTGTGAATTTTTTTACCTTATGCGCGTTAA TCATATTAATCAACTGCTGT CCGGGGCGAAACAGCTGTACCG CAACTTTTTTGTTGGGAAGGCGGGCCTCTTCTTTTTGCTATTACGCGGGGGA ACTCCTTTTTAACAGGTCAGTACCTTTAATTTTTTTGCTCCTTTTGTTTAAA GCGATGAGAGGATTACGGTG GGTGTTTTTTCTGGAAGTTTCAGTTGATTCCTTTTTCAATTCTGCGTAGTTT CGCCAATATAACATTCCTTCAG TTTCATTTGGTATGCAACTA AAGAATTTTTCCGGATATTCATCATAAGGGATTTTTACCGAACTGAAAGAGG AGCCGTTTTTGAACGAGGCGTCAACGTAACATTTTTAAGCTGCTCACACCAA ATTACCCAAACAGACGGTCA AGCTGTTTTTAAAAGGTGGCGGCAAAGAATTTTTTTAGCAAAATTAGACCCT AATAAATCCTAATAGTAGTAGCATTGTCGAAA TCCGCGACATCGCCTGATAAATTGTAACATCC CTGCTCCATGGATTTGTATC ATACAGGCAAATCAATTCTA Classification: DNA / Structure: SINGLE STRANDED / Synthetic?: Yes |
|---|---|
| Source (natural) | Organism: synthetic construct (others) |
| Molecular weight | Experimental: 1.03 MDa / Theoretical: 1.03 MDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.54 mg/mL |
|---|---|
| Buffer | pH: 8 / Details: 40 mM Tris-HCl, 20 mM acetic acid, 2 mM EDTA |
| Grid | Details: glow-discharged 200-mesh R1.2/1.3 Quantifoil grid |
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 100 % / Instrument: FEI VITROBOT MARK IV / Method: Blot for 1.5 second before plunging |
- Electron microscopy
Electron microscopy
| Microscope | JEOL 2200FS |
|---|---|
| Alignment procedure | Legacy - Astigmatism: Objective lens astigmatism was corrected at 80,000 times magnification |
| Specialist optics | Energy filter - Name: JEOL |
| Date | Apr 7, 2016 |
| Image recording | Category: CCD / Film or detector model: GATAN ULTRASCAN 4000 (4k x 4k) / Number real images: 152 / Average electron dose: 60 e/Å2 |
| Electron beam | Acceleration voltage: 200 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2 mm / Nominal defocus max: 0.004 µm / Nominal defocus min: 0.0015 µm / Nominal magnification: 40000 |
| Sample stage | Specimen holder model: GATAN LIQUID NITROGEN |
- Image processing
Image processing
| Details | The particles were selected manually using EMAN2 |
|---|---|
| CTF correction | Details: each particle |
| Final reconstruction | Applied symmetry - Point group: O (octahedral) / Algorithm: OTHER / Resolution.type: BY AUTHOR / Resolution: 40.0 Å / Resolution method: OTHER / Software - Name: EMAN2 / Number images used: 2008 |
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)





















