+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Cryo-EM structure of human Alpha-fetoprotein | |||||||||
 マップデータ マップデータ | ||||||||||
 試料 試料 |
| |||||||||
 キーワード キーワード | metal binding / fatty acids binding / METAL BINDING PROTEIN | |||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報progesterone metabolic process / ovulation from ovarian follicle / homeostasis of number of cells / Post-translational protein phosphorylation / Regulation of Insulin-like Growth Factor (IGF) transport and uptake by Insulin-like Growth Factor Binding Proteins (IGFBPs) / immune response / endoplasmic reticulum lumen / apoptotic process / extracellular space / metal ion binding / cytoplasm 類似検索 - 分子機能 | |||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 2.6 Å | |||||||||
 データ登録者 データ登録者 | Liu N / Liu K / Wu C / Liu Z / Li M / Wang J / Wang HW | |||||||||
| 資金援助 | 1件
| |||||||||
 引用 引用 |  ジャーナル: Nat Methods / 年: 2023 ジャーナル: Nat Methods / 年: 2023タイトル: Uniform thin ice on ultraflat graphene for high-resolution cryo-EM. 著者: Liming Zheng / Nan Liu / Xiaoyin Gao / Wenqing Zhu / Kun Liu / Cang Wu / Rui Yan / Jincan Zhang / Xin Gao / Yating Yao / Bing Deng / Jie Xu / Ye Lu / Zhongmin Liu / Mengsen Li / Xiaoding Wei ...著者: Liming Zheng / Nan Liu / Xiaoyin Gao / Wenqing Zhu / Kun Liu / Cang Wu / Rui Yan / Jincan Zhang / Xin Gao / Yating Yao / Bing Deng / Jie Xu / Ye Lu / Zhongmin Liu / Mengsen Li / Xiaoding Wei / Hong-Wei Wang / Hailin Peng /  要旨: Cryo-electron microscopy (cryo-EM) visualizes the atomic structure of macromolecules that are embedded in vitrified thin ice at their close-to-native state. However, the homogeneity of ice thickness, ...Cryo-electron microscopy (cryo-EM) visualizes the atomic structure of macromolecules that are embedded in vitrified thin ice at their close-to-native state. However, the homogeneity of ice thickness, a key factor to ensure high image quality, is poorly controlled during specimen preparation and has become one of the main challenges for high-resolution cryo-EM. Here we found that the uniformity of thin ice relies on the surface flatness of the supporting film, and developed a method to use ultraflat graphene (UFG) as the support for cryo-EM specimen preparation to achieve better control of vitreous ice thickness. We show that the uniform thin ice on UFG improves the image quality of vitrified specimens. Using such a method we successfully determined the three-dimensional structures of hemoglobin (64 kDa), α-fetoprotein (67 kDa) with no symmetry, and streptavidin (52 kDa) at a resolution of 3.5 Å, 2.6 Å and 2.2 Å, respectively. Furthermore, our results demonstrate the potential of UFG for the fields of cryo-electron tomography and structure-based drug discovery. | |||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_33861.map.gz emd_33861.map.gz | 12.1 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-33861-v30.xml emd-33861-v30.xml emd-33861.xml emd-33861.xml | 14.7 KB 14.7 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_33861.png emd_33861.png | 80.8 KB | ||
| マスクデータ |  emd_33861_msk_1.map emd_33861_msk_1.map | 12.9 MB |  マスクマップ マスクマップ | |
| Filedesc metadata |  emd-33861.cif.gz emd-33861.cif.gz | 5.4 KB | ||
| その他 |  emd_33861_half_map_1.map.gz emd_33861_half_map_1.map.gz emd_33861_half_map_2.map.gz emd_33861_half_map_2.map.gz | 11.9 MB 11.9 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33861 http://ftp.pdbj.org/pub/emdb/structures/EMD-33861 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33861 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33861 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_33861_validation.pdf.gz emd_33861_validation.pdf.gz | 759.4 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_33861_full_validation.pdf.gz emd_33861_full_validation.pdf.gz | 758.9 KB | 表示 | |
| XML形式データ |  emd_33861_validation.xml.gz emd_33861_validation.xml.gz | 9.2 KB | 表示 | |
| CIF形式データ |  emd_33861_validation.cif.gz emd_33861_validation.cif.gz | 10.8 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33861 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33861 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33861 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33861 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_33861.map.gz / 形式: CCP4 / 大きさ: 12.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_33861.map.gz / 形式: CCP4 / 大きさ: 12.9 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.0382 Å | ||||||||||||||||||||||||||||||||||||
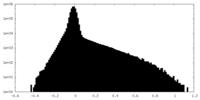
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-マスク #1
| ファイル |  emd_33861_msk_1.map emd_33861_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
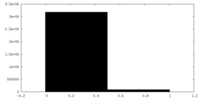
| 密度ヒストグラム |
-ハーフマップ: #1
| ファイル | emd_33861_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
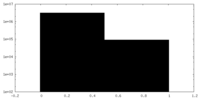
| 密度ヒストグラム |
-ハーフマップ: #2
| ファイル | emd_33861_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
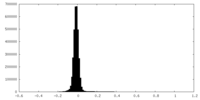
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Alpha-fetoprotein with no symmetry
| 全体 | 名称: Alpha-fetoprotein with no symmetry |
|---|---|
| 要素 |
|
-超分子 #1: Alpha-fetoprotein with no symmetry
| 超分子 | 名称: Alpha-fetoprotein with no symmetry / タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: all |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 67 KDa |
-分子 #1: Alpha-fetoprotein
| 分子 | 名称: Alpha-fetoprotein / タイプ: protein_or_peptide / ID: 1 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 68.757406 KDa |
| 組換発現 | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 配列 | 文字列: MKWVESIFLI FLLNFTESRT LHRNEYGIAS ILDSYQCTAE ISLADLATIF FAQFVQEATY KEVSKMVKDA LTAIEKPTGD EQSSGCLEN QLPAFLEELC HEKEILEKYG HSDCCSQSEE GRHNCFLAHK KPTPASIPLF QVPEPVTSCE AYEEDRETFM N KFIYEIAR ...文字列: MKWVESIFLI FLLNFTESRT LHRNEYGIAS ILDSYQCTAE ISLADLATIF FAQFVQEATY KEVSKMVKDA LTAIEKPTGD EQSSGCLEN QLPAFLEELC HEKEILEKYG HSDCCSQSEE GRHNCFLAHK KPTPASIPLF QVPEPVTSCE AYEEDRETFM N KFIYEIAR RHPFLYAPTI LLWAARYDKI IPSCCKAENA VECFQTKAAT VTKELRESSL LNQHACAVMK NFGTRTFQAI TV TKLSQKF TKVNFTEIQK LVLDVAHVHE HCCRGDVLDC LQDGEKIMSY ICSQQDTLSN KITECCKLTT LERGQCIIHA END EKPEGL SPNLNRFLGD RDFNQFSSGE KNIFLASFVH EYSRRHPQLA VSVILRVAKG YQELLEKCFQ TENPLECQDK GEEE LQKYI QESQALAKRS CGLFQKLGEY YLQNAFLVAY TKKAPQLTSS ELMAITRKMA ATAATCCQLS EDKLLACGEG AADII IGHL CIRHEMTPVN PGVGQCCTSS YANRRPCFSS LVVDETYVPP AFSDDKFIFH KDLCQAQGVA LQTMKQEFLI NLVKQK PQI TEEQLEAVIA DFSGLLEKCC QGQEQEVCFA EEGQKLISKT RAALGV UniProtKB: Alpha-fetoprotein |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 緩衝液 | pH: 7.4 |
|---|---|
| グリッド | モデル: Quantifoil R1.2/1.3 / 材質: GOLD / メッシュ: 300 / 支持フィルム - 材質: GRAPHENE / 支持フィルム - トポロジー: HOLEY ARRAY |
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 (6k x 4k) / 平均電子線量: 50.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD / 最大 デフォーカス(公称値): 1.2 µm / 最小 デフォーカス(公称値): 0.6 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
- 画像解析
画像解析
| 初期モデル | モデルのタイプ: PDB ENTRY PDBモデル - PDB ID: |
|---|---|
| 最終 再構成 | 解像度のタイプ: BY AUTHOR / 解像度: 2.6 Å / 解像度の算出法: FSC 0.143 CUT-OFF / 使用した粒子像数: 354264 |
| 初期 角度割当 | タイプ: MAXIMUM LIKELIHOOD |
| 最終 角度割当 | タイプ: MAXIMUM LIKELIHOOD |
 ムービー
ムービー コントローラー
コントローラー












 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)