[English] 日本語
 Yorodumi
Yorodumi- EMDB-33304: CryoEM structure of somatostatin receptor 4 (SSTR4) with Gi1 and ... -
+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Title | CryoEM structure of somatostatin receptor 4 (SSTR4) with Gi1 and J-2156 | |||||||||||||||||||||||||||
 Map data Map data | ||||||||||||||||||||||||||||
 Sample Sample |
| |||||||||||||||||||||||||||
| Function / homology |  Function and homology information Function and homology informationsomatostatin receptor activity / positive regulation of arachidonic acid secretion / negative regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway / neuropeptide binding / cellular response to glucocorticoid stimulus / plasma membrane => GO:0005886 / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / Adenylate cyclase inhibitory pathway / neuropeptide signaling pathway / positive regulation of protein localization to cell cortex ...somatostatin receptor activity / positive regulation of arachidonic acid secretion / negative regulation of adenylate cyclase-activating G protein-coupled receptor signaling pathway / neuropeptide binding / cellular response to glucocorticoid stimulus / plasma membrane => GO:0005886 / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / Adenylate cyclase inhibitory pathway / neuropeptide signaling pathway / positive regulation of protein localization to cell cortex / regulation of cAMP-mediated signaling / D2 dopamine receptor binding / G protein-coupled serotonin receptor binding / regulation of mitotic spindle organization / cellular response to forskolin / forebrain development / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / Peptide ligand-binding receptors / Regulation of insulin secretion / G protein-coupled receptor activity / G protein-coupled receptor binding / peptide binding / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G-protein activation / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through CDC42 / ADP signalling through P2Y purinoceptor 12 / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / Sensory perception of sweet, bitter, and umami (glutamate) taste / response to peptide hormone / photoreceptor disc membrane / Adrenaline,noradrenaline inhibits insulin secretion / Glucagon-type ligand receptors / Vasopressin regulates renal water homeostasis via Aquaporins / G alpha (z) signalling events / cellular response to catecholamine stimulus / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / ADORA2B mediated anti-inflammatory cytokines production / sensory perception of taste / ADP signalling through P2Y purinoceptor 1 / adenylate cyclase-activating dopamine receptor signaling pathway / G beta:gamma signalling through PI3Kgamma / cellular response to prostaglandin E stimulus / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / GPER1 signaling / GDP binding / G-protein beta-subunit binding / Inactivation, recovery and regulation of the phototransduction cascade / heterotrimeric G-protein complex / G alpha (12/13) signalling events / extracellular vesicle / signaling receptor complex adaptor activity / cell migration / Thrombin signalling through proteinase activated receptors (PARs) / GTPase binding / retina development in camera-type eye / phospholipase C-activating G protein-coupled receptor signaling pathway / Ca2+ pathway / cell cortex / midbody / G alpha (i) signalling events / fibroblast proliferation / G alpha (s) signalling events / G alpha (q) signalling events / cell population proliferation / Ras protein signal transduction / Extra-nuclear estrogen signaling / positive regulation of ERK1 and ERK2 cascade / neuron projection / G protein-coupled receptor signaling pathway / cell cycle / negative regulation of cell population proliferation / cell division / lysosomal membrane / GTPase activity / centrosome / synapse / protein-containing complex binding / nucleolus / GTP binding / magnesium ion binding / signal transduction / extracellular exosome / nucleoplasm / membrane / plasma membrane / cytoplasm / cytosol Similarity search - Function | |||||||||||||||||||||||||||
| Biological species |  Homo sapiens (human) / Homo sapiens (human) /  | |||||||||||||||||||||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.8 Å | |||||||||||||||||||||||||||
 Authors Authors | Wenli Z / Shuo H / Na Q / Wenbo Z / Mengjie L / Dehua Y / Ming-Wei W / Wu B / Zhao Q | |||||||||||||||||||||||||||
| Funding support |  China, 8 items China, 8 items
| |||||||||||||||||||||||||||
 Citation Citation |  Journal: Cell Res / Year: 2022 Journal: Cell Res / Year: 2022Title: Structural insights into ligand recognition and selectivity of somatostatin receptors. Authors: Wenli Zhao / Shuo Han / Na Qiu / Wenbo Feng / Mengjie Lu / Wenru Zhang / Mu Wang / Qingtong Zhou / Shutian Chen / Wei Xu / Juan Du / Xiaojing Chu / Cuiying Yi / Antao Dai / Liaoyuan Hu / ...Authors: Wenli Zhao / Shuo Han / Na Qiu / Wenbo Feng / Mengjie Lu / Wenru Zhang / Mu Wang / Qingtong Zhou / Shutian Chen / Wei Xu / Juan Du / Xiaojing Chu / Cuiying Yi / Antao Dai / Liaoyuan Hu / Michelle Y Shen / Yaping Sun / Qing Zhang / Yingli Ma / Wenge Zhong / Dehua Yang / Ming-Wei Wang / Beili Wu / Qiang Zhao /  Abstract: Somatostatin receptors (SSTRs) play versatile roles in inhibiting the secretion of multiple hormones such as growth hormone and thyroid-stimulating hormone, and thus are considered as targets for ...Somatostatin receptors (SSTRs) play versatile roles in inhibiting the secretion of multiple hormones such as growth hormone and thyroid-stimulating hormone, and thus are considered as targets for treating multiple tumors. Despite great progress made in therapeutic development against this diverse receptor family, drugs that target SSTRs still show limited efficacy with preferential binding affinity and conspicuous side-effects. Here, we report five structures of SSTR2 and SSTR4 in different states, including two crystal structures of SSTR2 in complex with a selective peptide antagonist and a non-peptide agonist, respectively, a cryo-electron microscopy (cryo-EM) structure of G-bound SSTR2 in the presence of the endogenous ligand SST-14, as well as two cryo-EM structures of G-bound SSTR4 in complex with SST-14 and a small-molecule agonist J-2156, respectively. By comparison of the SSTR structures in different states, molecular mechanisms of agonism and antagonism were illustrated. Together with computational and functional analyses, the key determinants responsible for ligand recognition and selectivity of different SSTR subtypes and multiform binding modes of peptide and non-peptide ligands were identified. Insights gained in this study will help uncover ligand selectivity of various SSTRs and accelerate the development of new molecules with better efficacy by targeting SSTRs. | |||||||||||||||||||||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_33304.map.gz emd_33304.map.gz | 59.7 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-33304-v30.xml emd-33304-v30.xml emd-33304.xml emd-33304.xml | 21.6 KB 21.6 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_33304.png emd_33304.png | 72.3 KB | ||
| Others |  emd_33304_half_map_1.map.gz emd_33304_half_map_1.map.gz emd_33304_half_map_2.map.gz emd_33304_half_map_2.map.gz | 49.7 MB 49.7 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33304 http://ftp.pdbj.org/pub/emdb/structures/EMD-33304 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33304 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33304 | HTTPS FTP |
-Validation report
| Summary document |  emd_33304_validation.pdf.gz emd_33304_validation.pdf.gz | 681.8 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_33304_full_validation.pdf.gz emd_33304_full_validation.pdf.gz | 681.4 KB | Display | |
| Data in XML |  emd_33304_validation.xml.gz emd_33304_validation.xml.gz | 12 KB | Display | |
| Data in CIF |  emd_33304_validation.cif.gz emd_33304_validation.cif.gz | 14.1 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33304 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33304 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33304 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33304 | HTTPS FTP |
-Related structure data
| Related structure data |  7xmtMC  7xmrC  7xmsC  7xn9C  7xnaC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_33304.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_33304.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Voxel size | X=Y=Z: 1.045 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: #2
| File | emd_33304_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
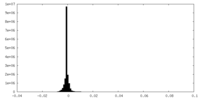
| Density Histograms |
-Half map: #1
| File | emd_33304_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
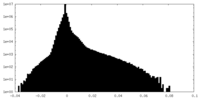
| Density Histograms |
- Sample components
Sample components
-Entire : Complex structure of somatostatin receptor 4 (SSTR4) with Gi and ...
| Entire | Name: Complex structure of somatostatin receptor 4 (SSTR4) with Gi and J-2156 |
|---|---|
| Components |
|
-Supramolecule #1: Complex structure of somatostatin receptor 4 (SSTR4) with Gi and ...
| Supramolecule | Name: Complex structure of somatostatin receptor 4 (SSTR4) with Gi and J-2156 type: complex / Chimera: Yes / ID: 1 / Parent: 0 / Macromolecule list: #1-#5 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:  Insecta environmental sample (insect) Insecta environmental sample (insect) |
-Macromolecule #1: Somatostatin receptor type 4
| Macromolecule | Name: Somatostatin receptor type 4 / type: protein_or_peptide / ID: 1 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.667375 KDa |
| Recombinant expression | Organism:  Insecta environmental sample (insect) Insecta environmental sample (insect) |
| Sequence | String: DYKDDDDGAP SAPSTLPPGG EEGLGTAWPS AANASSAPAE AEEAVAGPGD ARAAGMVAIQ CIYALVCLVG LVGNALVIFV ILRYAKMKT ATNIYLLNLA VADELFMLSV PFVASSAALR HWPFGSVLCR AVLSVDGLNM FTSVFCLTVL SVDRYVAVVH P LRAATYRR ...String: DYKDDDDGAP SAPSTLPPGG EEGLGTAWPS AANASSAPAE AEEAVAGPGD ARAAGMVAIQ CIYALVCLVG LVGNALVIFV ILRYAKMKT ATNIYLLNLA VADELFMLSV PFVASSAALR HWPFGSVLCR AVLSVDGLNM FTSVFCLTVL SVDRYVAVVH P LRAATYRR PSVAKLINLG VWLASLLVTL PIAIFADTRP ARGGQAVACN LQWPHPAWSA VFVVYTFLLG FLLPVLAIGL CY LLIVGKM RAVALRAGWQ QRRRSEKKIT RLVLMFVVVF VLCWMPFYVV QLLNLFVTSL DATVNHVSLI LSYANSCANP ILY GFLSDN FRRFFQRVLC LEFLEVLFQG PWSHPQFEKG GGSGGGSGGS AWSHPQFEK |
-Macromolecule #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(i) subunit alpha-1 type: protein_or_peptide / ID: 2 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 40.447141 KDa |
| Recombinant expression | Organism:  Insecta environmental sample (insect) Insecta environmental sample (insect) |
| Sequence | String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKCTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...String: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKCTIV KQMKIIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVTAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHASM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF |
-Macromolecule #3: single chain variable fragment of antibody
| Macromolecule | Name: single chain variable fragment of antibody / type: protein_or_peptide / ID: 3 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  |
| Molecular weight | Theoretical: 31.549107 KDa |
| Recombinant expression | Organism:  Insecta environmental sample (insect) Insecta environmental sample (insect) |
| Sequence | String: MKTIIALSYI FCLVFADYKD DDDGAPSEPD VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS SGGGGSGGGG S GGGGSDIV ...String: MKTIIALSYI FCLVFADYKD DDDGAPSEPD VQLVESGGGL VQPGGSRKLS CSASGFAFSS FGMHWVRQAP EKGLEWVAYI SSGSGTIYY ADTVKGRFTI SRDDPKNTLF LQMTSLRSED TAMYYCVRSI YYYGSSPFDF WGQGTTLTVS SGGGGSGGGG S GGGGSDIV MTQATSSVPV TPGESVSISC RSSKSLLHSN GNTYLYWFLQ RPGQSPQLLI YRMSNLASGV PDRFSGSGSG TA FTLTISR LEAEDVGVYY CMQHLEYPLT FGAGTKLELE FLEVLFQGPH HHHHH |
-Macromolecule #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 type: protein_or_peptide / ID: 4 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 7.861143 KDa |
| Recombinant expression | Organism:  Insecta environmental sample (insect) Insecta environmental sample (insect) |
| Sequence | String: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L |
-Macromolecule #5: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| Macromolecule | Name: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 type: protein_or_peptide / ID: 5 / Number of copies: 1 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 38.744371 KDa |
| Recombinant expression | Organism:  Insecta environmental sample (insect) Insecta environmental sample (insect) |
| Sequence | String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI ...String: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI VTSSGDTTCA LWDIETGQQT TTFTGHTGDV MSLSLAPDTR LFVSGACDAS AKLWDVREGM CRQTFTGHES DI NAICFFP NGNAFATGSD DATCRLFDLR ADQELMTYSH DNIICGITSV SFSKSGRLLL AGYDDFNCNV WDALKADRAG VLA GHDNRV SCLGVTDDGM AVATGSWDSF LKIWN |
-Macromolecule #6: (2~{S})-2-[[(2~{S})-4-azanyl-2-[(4-methylnaphthalen-1-yl)sulfonyl...
| Macromolecule | Name: (2~{S})-2-[[(2~{S})-4-azanyl-2-[(4-methylnaphthalen-1-yl)sulfonylamino]butanoyl]amino]-3-phenyl-propanimidic acid type: ligand / ID: 6 / Number of copies: 1 / Formula: I8B |
|---|---|
| Molecular weight | Theoretical: 468.569 Da |
| Chemical component information | 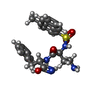 ChemComp-I8B: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Material: NICKEL/TITANIUM |
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Average electron dose: 70.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 1.3 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.8 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 600908 |
|---|---|
| Initial angle assignment | Type: MAXIMUM LIKELIHOOD |
| Final angle assignment | Type: MAXIMUM LIKELIHOOD |
 Movie
Movie Controller
Controller



































 Z
Z Y
Y X
X