+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 |  | |||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | CryoEM structure of the somatostatin receptor 2 (SSTR2) in complex with Gi1 and its endogeneous peptide ligand SST-14 | |||||||||||||||||||||||||||
 マップデータ マップデータ | ||||||||||||||||||||||||||||
 試料 試料 |
| |||||||||||||||||||||||||||
 キーワード キーワード | G protein-coupled receptor / somatostatin receptor 2 / cryo-EM / SIGNALING PROTEIN | |||||||||||||||||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報somatostatin signaling pathway / MECP2 regulates transcription of neuronal ligands / somatostatin receptor activity / hormone-mediated apoptotic signaling pathway / response to acidic pH / neuropeptide binding / response to steroid hormone / hyperosmotic response / cellular response to glucocorticoid stimulus / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger ...somatostatin signaling pathway / MECP2 regulates transcription of neuronal ligands / somatostatin receptor activity / hormone-mediated apoptotic signaling pathway / response to acidic pH / neuropeptide binding / response to steroid hormone / hyperosmotic response / cellular response to glucocorticoid stimulus / G protein-coupled receptor signaling pathway, coupled to cyclic nucleotide second messenger / response to amino acid / neuropeptide signaling pathway / neuronal dense core vesicle / regulation of postsynaptic membrane neurotransmitter receptor levels / adenylate cyclase inhibitor activity / digestion / positive regulation of protein localization to cell cortex / T cell migration / Adenylate cyclase inhibitory pathway / D2 dopamine receptor binding / response to prostaglandin E / adenylate cyclase regulator activity / G protein-coupled serotonin receptor binding / adenylate cyclase-inhibiting serotonin receptor signaling pathway / response to nutrient / cellular response to forskolin / regulation of cell migration / regulation of mitotic spindle organization / Peptide ligand-binding receptors / PDZ domain binding / Regulation of insulin secretion / cellular response to estradiol stimulus / positive regulation of cholesterol biosynthetic process / negative regulation of insulin secretion / G protein-coupled receptor binding / hormone activity / response to peptide hormone / adenylate cyclase-inhibiting G protein-coupled receptor signaling pathway / GABA-ergic synapse / centriolar satellite / adenylate cyclase-modulating G protein-coupled receptor signaling pathway / G-protein beta/gamma-subunit complex binding / Olfactory Signaling Pathway / Activation of the phototransduction cascade / G beta:gamma signalling through PLC beta / Presynaptic function of Kainate receptors / Thromboxane signalling through TP receptor / G protein-coupled acetylcholine receptor signaling pathway / Activation of G protein gated Potassium channels / Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits / G-protein activation / G beta:gamma signalling through CDC42 / Prostacyclin signalling through prostacyclin receptor / Glucagon signaling in metabolic regulation / G beta:gamma signalling through BTK / Synthesis, secretion, and inactivation of Glucagon-like Peptide-1 (GLP-1) / ADP signalling through P2Y purinoceptor 12 / photoreceptor disc membrane / Sensory perception of sweet, bitter, and umami (glutamate) taste / Glucagon-type ligand receptors / Adrenaline,noradrenaline inhibits insulin secretion / GDP binding / Vasopressin regulates renal water homeostasis via Aquaporins / Glucagon-like Peptide-1 (GLP1) regulates insulin secretion / G alpha (z) signalling events / ADP signalling through P2Y purinoceptor 1 / cellular response to catecholamine stimulus / ADORA2B mediated anti-inflammatory cytokines production / G beta:gamma signalling through PI3Kgamma / Cooperation of PDCL (PhLP1) and TRiC/CCT in G-protein beta folding / adenylate cyclase-activating dopamine receptor signaling pathway / cell-cell signaling / GPER1 signaling / Inactivation, recovery and regulation of the phototransduction cascade / G-protein beta-subunit binding / cellular response to prostaglandin E stimulus / heterotrimeric G-protein complex / G alpha (12/13) signalling events / sensory perception of taste / extracellular vesicle / signaling receptor complex adaptor activity / Thrombin signalling through proteinase activated receptors (PARs) / retina development in camera-type eye / G protein activity / GTPase binding / Ca2+ pathway / fibroblast proliferation / midbody / cell cortex / High laminar flow shear stress activates signaling by PIEZO1 and PECAM1:CDH5:KDR in endothelial cells / G alpha (i) signalling events / G alpha (s) signalling events / phospholipase C-activating G protein-coupled receptor signaling pathway / G alpha (q) signalling events / chemical synaptic transmission / 加水分解酵素; 酸無水物に作用; GTPに作用・細胞または細胞小器官の運動に関与 / Ras protein signal transduction / cell surface receptor signaling pathway / Extra-nuclear estrogen signaling / cell population proliferation 類似検索 - 分子機能 | |||||||||||||||||||||||||||
| 生物種 |  Homo sapiens (ヒト) Homo sapiens (ヒト) | |||||||||||||||||||||||||||
| 手法 | 単粒子再構成法 / クライオ電子顕微鏡法 / 解像度: 3.1 Å | |||||||||||||||||||||||||||
 データ登録者 データ登録者 | Wenli Z / Shuo H / Na Q / Wenbo Z / Mengjie L / Dehua Y / Ming-Wei W / Wu B / Zhao Q | |||||||||||||||||||||||||||
| 資金援助 |  中国, 8件 中国, 8件
| |||||||||||||||||||||||||||
 引用 引用 |  ジャーナル: Cell Res / 年: 2022 ジャーナル: Cell Res / 年: 2022タイトル: Structural insights into ligand recognition and selectivity of somatostatin receptors. 著者: Wenli Zhao / Shuo Han / Na Qiu / Wenbo Feng / Mengjie Lu / Wenru Zhang / Mu Wang / Qingtong Zhou / Shutian Chen / Wei Xu / Juan Du / Xiaojing Chu / Cuiying Yi / Antao Dai / Liaoyuan Hu / ...著者: Wenli Zhao / Shuo Han / Na Qiu / Wenbo Feng / Mengjie Lu / Wenru Zhang / Mu Wang / Qingtong Zhou / Shutian Chen / Wei Xu / Juan Du / Xiaojing Chu / Cuiying Yi / Antao Dai / Liaoyuan Hu / Michelle Y Shen / Yaping Sun / Qing Zhang / Yingli Ma / Wenge Zhong / Dehua Yang / Ming-Wei Wang / Beili Wu / Qiang Zhao /  要旨: Somatostatin receptors (SSTRs) play versatile roles in inhibiting the secretion of multiple hormones such as growth hormone and thyroid-stimulating hormone, and thus are considered as targets for ...Somatostatin receptors (SSTRs) play versatile roles in inhibiting the secretion of multiple hormones such as growth hormone and thyroid-stimulating hormone, and thus are considered as targets for treating multiple tumors. Despite great progress made in therapeutic development against this diverse receptor family, drugs that target SSTRs still show limited efficacy with preferential binding affinity and conspicuous side-effects. Here, we report five structures of SSTR2 and SSTR4 in different states, including two crystal structures of SSTR2 in complex with a selective peptide antagonist and a non-peptide agonist, respectively, a cryo-electron microscopy (cryo-EM) structure of G-bound SSTR2 in the presence of the endogenous ligand SST-14, as well as two cryo-EM structures of G-bound SSTR4 in complex with SST-14 and a small-molecule agonist J-2156, respectively. By comparison of the SSTR structures in different states, molecular mechanisms of agonism and antagonism were illustrated. Together with computational and functional analyses, the key determinants responsible for ligand recognition and selectivity of different SSTR subtypes and multiform binding modes of peptide and non-peptide ligands were identified. Insights gained in this study will help uncover ligand selectivity of various SSTRs and accelerate the development of new molecules with better efficacy by targeting SSTRs. | |||||||||||||||||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| 添付画像 |
|---|
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_33302.map.gz emd_33302.map.gz | 59.6 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-33302-v30.xml emd-33302-v30.xml emd-33302.xml emd-33302.xml | 21 KB 21 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| 画像 |  emd_33302.png emd_33302.png | 82.7 KB | ||
| Filedesc metadata |  emd-33302.cif.gz emd-33302.cif.gz | 6.3 KB | ||
| その他 |  emd_33302_half_map_1.map.gz emd_33302_half_map_1.map.gz emd_33302_half_map_2.map.gz emd_33302_half_map_2.map.gz | 49.5 MB 49.5 MB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-33302 http://ftp.pdbj.org/pub/emdb/structures/EMD-33302 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33302 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-33302 | HTTPS FTP |
-検証レポート
| 文書・要旨 |  emd_33302_validation.pdf.gz emd_33302_validation.pdf.gz | 798.2 KB | 表示 |  EMDB検証レポート EMDB検証レポート |
|---|---|---|---|---|
| 文書・詳細版 |  emd_33302_full_validation.pdf.gz emd_33302_full_validation.pdf.gz | 797.8 KB | 表示 | |
| XML形式データ |  emd_33302_validation.xml.gz emd_33302_validation.xml.gz | 12.1 KB | 表示 | |
| CIF形式データ |  emd_33302_validation.cif.gz emd_33302_validation.cif.gz | 14.3 KB | 表示 | |
| アーカイブディレクトリ |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33302 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33302 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33302 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-33302 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_33302.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_33302.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 | 画像のコントロール
画像は Spider により作成 | ||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.045 Å | ||||||||||||||||||||||||||||||||||||
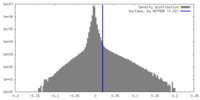
| 密度 |
| ||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
|
-添付データ
-ハーフマップ: #1
| ファイル | emd_33302_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
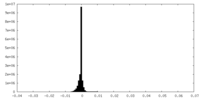
| 密度ヒストグラム |
-ハーフマップ: #2
| ファイル | emd_33302_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 投影像・断面図 |
| ||||||||||||
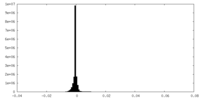
| 密度ヒストグラム |
- 試料の構成要素
試料の構成要素
-全体 : Complex structure of the somatostatin receptor 2 (SSTR2) in compl...
| 全体 | 名称: Complex structure of the somatostatin receptor 2 (SSTR2) in complex with Gi1 and its endogeneous peptide ligand SST-14 |
|---|---|
| 要素 |
|
-超分子 #1: Complex structure of the somatostatin receptor 2 (SSTR2) in compl...
| 超分子 | 名称: Complex structure of the somatostatin receptor 2 (SSTR2) in complex with Gi1 and its endogeneous peptide ligand SST-14 タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: all |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
-分子 #1: Somatostatin receptor type 2
| 分子 | 名称: Somatostatin receptor type 2 / タイプ: protein_or_peptide / ID: 1 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 45.308605 KDa |
| 組換発現 | 生物種:  Insecta environmental sample (昆虫) Insecta environmental sample (昆虫) |
| 配列 | 文字列: DYKDDDDGAP DMADEPLNGS HTWLSIPFDL NGSVVSTNTS NQTEPYYDLT SNAVLTFIYF VVCIIGLCGN TLVIYVILRY AKMKTITNI YILNLAIADE LFMLGLPFLA MQVALVHWPF GKAICRVVMT VDGINQFTSI FCLTVMSIDR YLAVVHPIKS A KWRRPRTA ...文字列: DYKDDDDGAP DMADEPLNGS HTWLSIPFDL NGSVVSTNTS NQTEPYYDLT SNAVLTFIYF VVCIIGLCGN TLVIYVILRY AKMKTITNI YILNLAIADE LFMLGLPFLA MQVALVHWPF GKAICRVVMT VDGINQFTSI FCLTVMSIDR YLAVVHPIKS A KWRRPRTA KMITMAVWGV SLLVILPIMI YAGLRSNQWG RSSCTINWPG ESGAWYTGFI IYTFILGFLV PLTIICLCYL FI IIKVKSS GIRVGSSKRK KSEKKVTRMV SIVVAVFIFC WLPFYIFNVS SVSMAISPTP ALKGMFDFVV VLTYANSCAN PIL YAFLSD NFKKSFQNVL CLVKVSGTDD GERSDSKQDK SRLNETTETQ RTEFLEVLFQ GPWSHPQFEK GGGSGGGSGG SAWS HPQFE K UniProtKB: Somatostatin receptor type 2 |
-分子 #2: Guanine nucleotide-binding protein G(i) subunit alpha-1
| 分子 | 名称: Guanine nucleotide-binding protein G(i) subunit alpha-1 タイプ: protein_or_peptide / ID: 2 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 40.435086 KDa |
| 組換発現 | 生物種:  Insecta environmental sample (昆虫) Insecta environmental sample (昆虫) |
| 配列 | 文字列: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKCTIV KQMKTIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI ...文字列: MGCTLSAEDK AAVERSKMID RNLREDGEKA AREVKLLLLG AGESGKCTIV KQMKTIHEAG YSEEECKQYK AVVYSNTIQS IIAIIRAMG RLKIDFGDSA RADDARQLFV LAGAAEEGFM TAELAGVIKR LWKDSGVQAC FNRSREYQLN DSAAYYLNDL D RIAQPNYI PTQQDVLRTR VKTTGIVETH FTFKDLHFKM FDVTAQRSER KKWIHCFEGV TAIIFCVALS DYDLVLAEDE EM NRMHASM KLFDSICNNK WFTDTSIILF LNKKDLFEEK IKKSPLTICY PEYAGSNTYE EAAAYIQCQF EDLNKRKDTK EIY THFTCS TDTKNVQFVF DAVTDVIIKN NLKDCGLF UniProtKB: Guanine nucleotide-binding protein G(i) subunit alpha-1 |
-分子 #3: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1
| 分子 | 名称: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 タイプ: protein_or_peptide / ID: 3 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 38.744371 KDa |
| 組換発現 | 生物種:  Insecta environmental sample (昆虫) Insecta environmental sample (昆虫) |
| 配列 | 文字列: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI ...文字列: MHHHHHHGSL LQSELDQLRQ EAEQLKNQIR DARKACADAT LSQITNNIDP VGRIQMRTRR TLRGHLAKIY AMHWGTDSRL LVSASQDGK LIIWDSYTTN KVHAIPLRSS WVMTCAYAPS GNYVACGGLD NICSIYNLKT REGNVRVSRE LAGHTGYLSC C RFLDDNQI VTSSGDTTCA LWDIETGQQT TTFTGHTGDV MSLSLAPDTR LFVSGACDAS AKLWDVREGM CRQTFTGHES DI NAICFFP NGNAFATGSD DATCRLFDLR ADQELMTYSH DNIICGITSV SFSKSGRLLL AGYDDFNCNV WDALKADRAG VLA GHDNRV SCLGVTDDGM AVATGSWDSF LKIWN UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1 |
-分子 #4: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2
| 分子 | 名称: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 タイプ: protein_or_peptide / ID: 4 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 7.861143 KDa |
| 組換発現 | 生物種:  Insecta environmental sample (昆虫) Insecta environmental sample (昆虫) |
| 配列 | 文字列: MASNNTASIA QARKLVEQLK MEANIDRIKV SKAAADLMAY CEAHAKEDPL LTPVPASENP FREKKFFCAI L UniProtKB: Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2 |
-分子 #5: Somatostatin-14
| 分子 | 名称: Somatostatin-14 / タイプ: protein_or_peptide / ID: 5 / コピー数: 1 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:  Homo sapiens (ヒト) Homo sapiens (ヒト) |
| 分子量 | 理論値: 1.641909 KDa |
| 配列 | 文字列: AGCKNFFWKT FTSC UniProtKB: Somatostatin |
-実験情報
-構造解析
| 手法 | クライオ電子顕微鏡法 |
|---|---|
 解析 解析 | 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 1.0 mg/mL |
|---|---|
| 緩衝液 | pH: 7.5 |
| グリッド | 材質: NICKEL/TITANIUM |
| 凍結 | 凍結剤: ETHANE |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 撮影 | フィルム・検出器のモデル: GATAN K3 BIOQUANTUM (6k x 4k) 平均電子線量: 70.0 e/Å2 |
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD 最大 デフォーカス(公称値): 2.3000000000000003 µm 最小 デフォーカス(公称値): 1.3 µm |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
- 画像解析
画像解析
| 初期モデル | モデルのタイプ: INSILICO MODEL |
|---|---|
| 最終 再構成 | 解像度のタイプ: BY AUTHOR / 解像度: 3.1 Å / 解像度の算出法: FSC 0.143 CUT-OFF / 使用した粒子像数: 696255 |
| 初期 角度割当 | タイプ: MAXIMUM LIKELIHOOD |
| 最終 角度割当 | タイプ: MAXIMUM LIKELIHOOD |
 ムービー
ムービー コントローラー
コントローラー









































 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)