+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
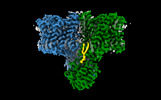
| Title | Cryo-EM structure of human BTR1 in the outward-facing state. | |||||||||
 Map data Map data | Sharpened map of wild-type BTR1 | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | SLC4A11 / BTR1 / SLC transporter / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationborate transport / borate efflux transmembrane transporter activity / active borate transmembrane transporter activity / regulation of mesenchymal stem cell differentiation / fluid transport / water transmembrane transporter activity / monoatomic ion homeostasis / intracellular monoatomic cation homeostasis / cellular hypotonic response / proton channel activity ...borate transport / borate efflux transmembrane transporter activity / active borate transmembrane transporter activity / regulation of mesenchymal stem cell differentiation / fluid transport / water transmembrane transporter activity / monoatomic ion homeostasis / intracellular monoatomic cation homeostasis / cellular hypotonic response / proton channel activity / symporter activity / solute:inorganic anion antiporter activity / vesicle membrane / sodium channel activity / bicarbonate transport / bicarbonate transmembrane transporter activity / monoatomic anion transport / sodium ion transport / proton transmembrane transporter activity / transmembrane transporter activity / proton transmembrane transport / regulation of mitochondrial membrane potential / transmembrane transport / cellular response to oxidative stress / basolateral plasma membrane / protein dimerization activity / apical plasma membrane / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method | single particle reconstruction / cryo EM / Resolution: 2.94 Å | |||||||||
 Authors Authors | Yin Y / Lu Y / Zuo P | |||||||||
| Funding support |  China, 2 items China, 2 items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2023 Journal: Nat Commun / Year: 2023Title: Structural insights into the conformational changes of BTR1/SLC4A11 in complex with PIP. Authors: Yishuo Lu / Peng Zuo / Hongyi Chen / Hui Shan / Weize Wang / Zonglin Dai / He Xu / Yayu Chen / Ling Liang / Dian Ding / Yan Jin / Yuxin Yin /  Abstract: BTR1 (SLC4A11) is a NH stimulated H (OH) transporter belonging to the SLC4 family. Dysfunction of BTR1 leads to diseases such as congenital hereditary endothelial dystrophy (CHED) and Fuchs ...BTR1 (SLC4A11) is a NH stimulated H (OH) transporter belonging to the SLC4 family. Dysfunction of BTR1 leads to diseases such as congenital hereditary endothelial dystrophy (CHED) and Fuchs endothelial corneal dystrophy (FECD). However, the mechanistic basis of BTR1 activation by alkaline pH, transport activity regulation and pathogenic mutations remains elusive. Here, we present cryo-EM structures of human BTR1 in the outward-facing state in complex with its activating ligands PIP and the inward-facing state with the pathogenic R125H mutation. We reveal that PIP binds at the interface between the transmembrane domain and the N-terminal cytosolic domain of BTR1. Disruption of either the PIP binding site or protonation of PIP phosphate groups by acidic pH can transform BTR1 into an inward-facing conformation. Our results provide insights into the mechanisms of how the transport activity and conformation changes of BTR1 are regulated by PIP binding and interaction of TMD and NTD. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_32942.map.gz emd_32942.map.gz | 230.2 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-32942-v30.xml emd-32942-v30.xml emd-32942.xml emd-32942.xml | 15.3 KB 15.3 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_32942.png emd_32942.png | 57 KB | ||
| Filedesc metadata |  emd-32942.cif.gz emd-32942.cif.gz | 6.1 KB | ||
| Others |  emd_32942_half_map_1.map.gz emd_32942_half_map_1.map.gz emd_32942_half_map_2.map.gz emd_32942_half_map_2.map.gz | 226.6 MB 226.6 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-32942 http://ftp.pdbj.org/pub/emdb/structures/EMD-32942 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32942 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-32942 | HTTPS FTP |
-Validation report
| Summary document |  emd_32942_validation.pdf.gz emd_32942_validation.pdf.gz | 984.2 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_32942_full_validation.pdf.gz emd_32942_full_validation.pdf.gz | 983.8 KB | Display | |
| Data in XML |  emd_32942_validation.xml.gz emd_32942_validation.xml.gz | 15.9 KB | Display | |
| Data in CIF |  emd_32942_validation.cif.gz emd_32942_validation.cif.gz | 18.8 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32942 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32942 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32942 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-32942 | HTTPS FTP |
-Related structure data
| Related structure data |  7x1iMC  7x1gC  7x1hC  7x1jC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_32942.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_32942.map.gz / Format: CCP4 / Size: 244.1 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Sharpened map of wild-type BTR1 | ||||||||||||||||||||
| Voxel size | X=Y=Z: 0.821 Å | ||||||||||||||||||||
| Density |
| ||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Half map: Half-A map of wild-type BTR1
| File | emd_32942_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-A map of wild-type BTR1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-B map of wild-type BTR1
| File | emd_32942_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-B map of wild-type BTR1 | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Solute carrier family 4 member 11, isoform B
| Entire | Name: Solute carrier family 4 member 11, isoform B |
|---|---|
| Components |
|
-Supramolecule #1: Solute carrier family 4 member 11, isoform B
| Supramolecule | Name: Solute carrier family 4 member 11, isoform B / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1 |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 199 KDa |
-Macromolecule #1: Isoform 1 of Solute carrier family 4 member 11
| Macromolecule | Name: Isoform 1 of Solute carrier family 4 member 11 / type: protein_or_peptide / ID: 1 / Number of copies: 2 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 99.684438 KDa |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
| Sequence | String: MSQVGGRGDR CTQEVQGLVH GAGDLSASLA ENSPTMSQNG YFEDSSYYKC DTDDTFEARE EILGDEAFDT ANSSIVSGES IRFFVNVNL EMQATNTENE ATSGGCVLLH TSRKYLKLKN FKEEIRAHRD LDGFLAQASI VLNETATSLD NVLRTMLRRF A RDPDNNEP ...String: MSQVGGRGDR CTQEVQGLVH GAGDLSASLA ENSPTMSQNG YFEDSSYYKC DTDDTFEARE EILGDEAFDT ANSSIVSGES IRFFVNVNL EMQATNTENE ATSGGCVLLH TSRKYLKLKN FKEEIRAHRD LDGFLAQASI VLNETATSLD NVLRTMLRRF A RDPDNNEP NCNLDLLMAM LFTDAGAPMR GKVHLLSDTI QGVTATVTGV RYQQSWLCII CTMKALQKRH VCISRLVRPQ NW GENSCEV RFVILVLAPP KMKSTKTAME VARTFATMFS DIAFRQKLLE TRTEEEFKEA LVHQRQLLTM VSHGPVAPRT KER STVSLP AHRHPEPPKC KDFVPFGKGI REDIARRFPL YPLDFTDGII GKNKAVGKYI TTTLFLYFAC LLPTIAFGSL NDEN TDGAI DVQKTIAGQS IGGLLYALFS GQPLVILLTT APLALYIQVI RVICDDYDLD FNSFYAWTGL WNSFFLALYA FFNLS LVMS LFKRSTEEII ALFISITFVL DAVKGTVKIF WKYYYGHYLD DYHTKRTSSL VSLSGLGASL NASLHTALNA SFLASP TEL PSATHSGQAT AVLSLLIMLG TLWLGYTLYQ FKKSPYLHPC VREILSDCAL PIAVLAFSLI SSHGFREIEM SKFRYNP SE SPFAMAQIQS LSLRAVSGAM GLGFLLSMLF FIEQNLVAAL VNAPENRLVK GTAYHWDLLL LAIINTGLSL FGLPWIHA A YPHSPLHVRA LALVEERVEN GHIYDTIVNV KETRLTSLGA SVLVGLSLLL LPVPLQWIPK PVLYGLFLYI ALTSLDGNQ LVQRVALLLK EQTAYPPTHY IRRVPQRKIH YFTGLQVLQL LLLCAFGMSS LPYMKMIFPL IMIAMIPIRY ILLPRIIEAK YLDVMDAEH RP UniProtKB: Solute carrier family 4 member 11 |
-Macromolecule #2: [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tr...
| Macromolecule | Name: [(2R)-1-octadecanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phospho ryl]oxy-propan-2-yl] (8Z)-icosa-5,8,11,14-tetraenoate type: ligand / ID: 2 / Number of copies: 2 / Formula: PT5 |
|---|---|
| Molecular weight | Theoretical: 1.047088 KDa |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Vitrification | Cryogen name: ETHANE |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: SUPER-RESOLUTION / Average electron dose: 50.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 1.8 µm / Nominal defocus min: 1.5 µm |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Startup model | Type of model: OTHER |
|---|---|
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 2.94 Å / Resolution method: FSC 0.143 CUT-OFF / Software - Name: cryoSPARC (ver. 3.1.0) / Number images used: 92740 |
| Initial angle assignment | Type: OTHER |
| Final angle assignment | Type: OTHER |
 Movie
Movie Controller
Controller






 Z
Z Y
Y X
X